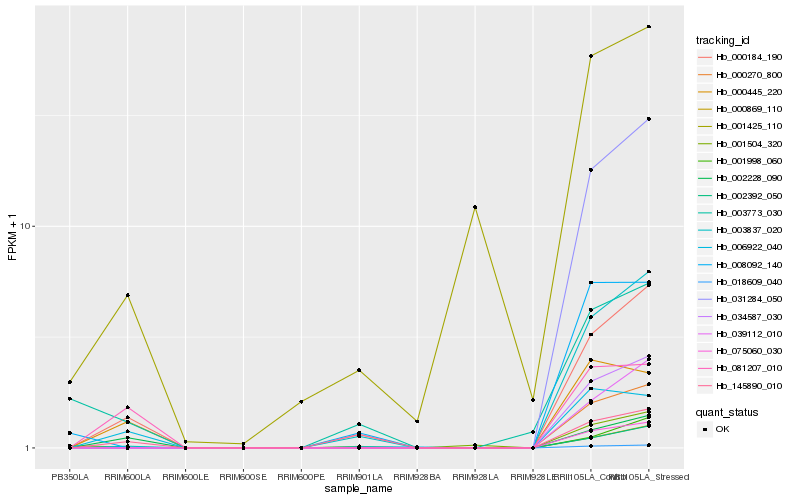
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000184_190 |
0.0 |
- |
- |
hypothetical protein VITISV_016970 [Vitis vinifera] |
| 2 |
Hb_003837_020 |
0.1466610127 |
- |
- |
PREDICTED: uncharacterized protein LOC103710465 isoform X1 [Phoenix dactylifera] |
| 3 |
Hb_002228_090 |
0.1766961264 |
- |
- |
hypothetical protein POPTR_0006s23560g [Populus trichocarpa] |
| 4 |
Hb_031284_050 |
0.1950015011 |
- |
- |
- |
| 5 |
Hb_002392_050 |
0.1965243451 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X4 [Jatropha curcas] |
| 6 |
Hb_000869_110 |
0.196559973 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_039112_010 |
0.1968665892 |
- |
- |
- |
| 8 |
Hb_034587_030 |
0.1977211252 |
- |
- |
- |
| 9 |
Hb_018609_040 |
0.1985380254 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 10 |
Hb_000270_800 |
0.1985667978 |
- |
- |
- |
| 11 |
Hb_075060_030 |
0.1985917118 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 12 |
Hb_006922_040 |
0.2012809957 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_145890_010 |
0.2095132675 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001504_320 |
0.2121020086 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 15 |
Hb_001998_060 |
0.2124351971 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_081207_010 |
0.2201894808 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 17 |
Hb_000445_220 |
0.2246240459 |
- |
- |
- |
| 18 |
Hb_008092_140 |
0.2318036137 |
- |
- |
PREDICTED: uncharacterized protein LOC103636948 [Zea mays] |
| 19 |
Hb_003773_030 |
0.2322089462 |
- |
- |
- |
| 20 |
Hb_001425_110 |
0.2364880874 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |