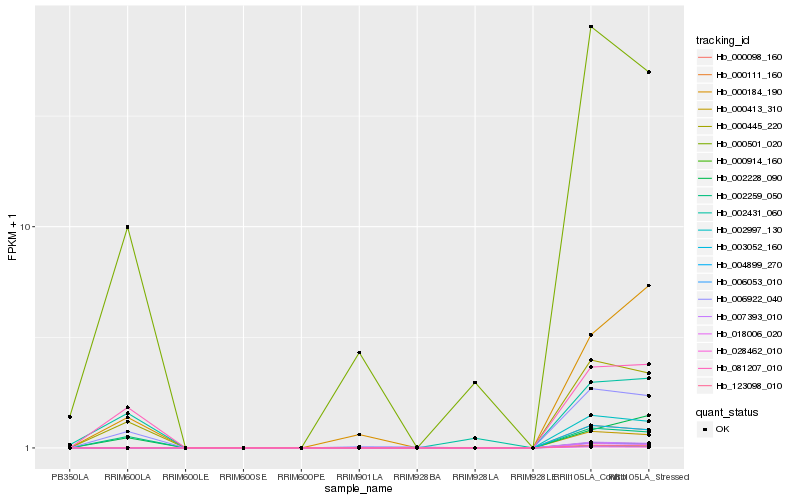
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_006922_040 |
0.0801119461 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_081207_010 |
0.0914254105 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 4 |
Hb_000501_020 |
0.1303553221 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 5 |
Hb_002259_050 |
0.1496461217 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002228_090 |
0.1885916688 |
- |
- |
hypothetical protein POPTR_0006s23560g [Populus trichocarpa] |
| 7 |
Hb_002431_060 |
0.1955447052 |
- |
- |
- |
| 8 |
Hb_000184_190 |
0.2246240459 |
- |
- |
hypothetical protein VITISV_016970 [Vitis vinifera] |
| 9 |
Hb_002997_130 |
0.2340407032 |
- |
- |
- |
| 10 |
Hb_028462_010 |
0.2340407283 |
- |
- |
PREDICTED: uncharacterized protein LOC101509514 [Cicer arietinum] |
| 11 |
Hb_123098_010 |
0.2340407305 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_004899_270 |
0.2340407481 |
- |
- |
PREDICTED: beta-galactosidase 15-like [Cucumis sativus] |
| 13 |
Hb_000111_160 |
0.2340407747 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 14 |
Hb_018006_020 |
0.2340408444 |
- |
- |
PREDICTED: uncharacterized protein LOC105641417 [Jatropha curcas] |
| 15 |
Hb_006053_010 |
0.2340408667 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 16 |
Hb_000914_160 |
0.2340408833 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Glycine max] |
| 17 |
Hb_007393_010 |
0.2340409102 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 18 |
Hb_000413_310 |
0.2340410722 |
- |
- |
- |
| 19 |
Hb_000098_160 |
0.2340410961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003052_160 |
0.2340410961 |
- |
- |
- |