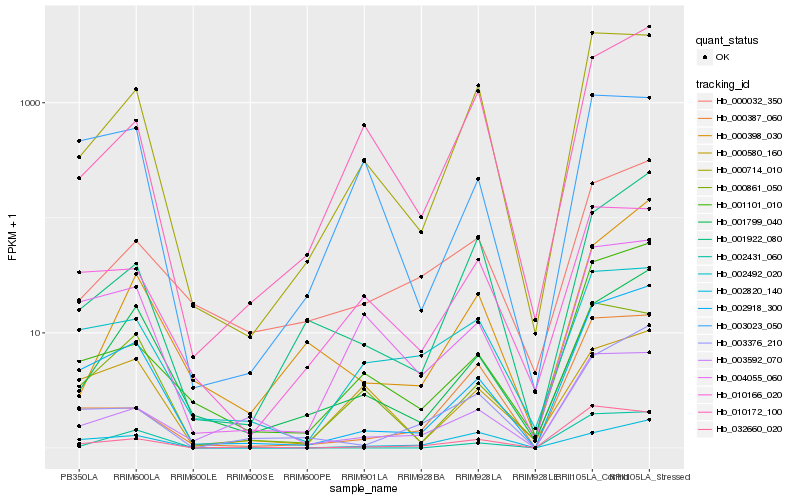
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002918_300 |
0.0 |
- |
- |
- |
| 2 |
Hb_001101_010 |
0.1585518429 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 3 |
Hb_000714_010 |
0.1681124659 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 4 |
Hb_001799_040 |
0.1708361926 |
- |
- |
- |
| 5 |
Hb_002431_060 |
0.1714974495 |
- |
- |
- |
| 6 |
Hb_003376_210 |
0.1743995856 |
- |
- |
- |
| 7 |
Hb_032660_020 |
0.1757125109 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 8 |
Hb_004055_060 |
0.1771133977 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 9 |
Hb_000861_050 |
0.1809081374 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 10 |
Hb_001922_080 |
0.1959612994 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 11 |
Hb_000387_060 |
0.2003947968 |
- |
- |
- |
| 12 |
Hb_002492_020 |
0.2035474965 |
- |
- |
PREDICTED: uncharacterized protein LOC104789613 [Camelina sativa] |
| 13 |
Hb_010166_020 |
0.2056377286 |
transcription factor |
TF Family: NF-YC |
ccaat-binding transcription factor, putative [Ricinus communis] |
| 14 |
Hb_003592_070 |
0.207668342 |
- |
- |
- |
| 15 |
Hb_000398_030 |
0.2126036304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003023_050 |
0.2154997513 |
- |
- |
hypothetical protein POPTR_0013s10320g [Populus trichocarpa] |
| 17 |
Hb_000032_350 |
0.2166681989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010172_100 |
0.218963952 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 19 |
Hb_002820_140 |
0.2220004705 |
- |
- |
PREDICTED: uncharacterized protein LOC105638912 [Jatropha curcas] |
| 20 |
Hb_000580_160 |
0.2221117099 |
- |
- |
Nitrate transporter 1.5 [Morus notabilis] |