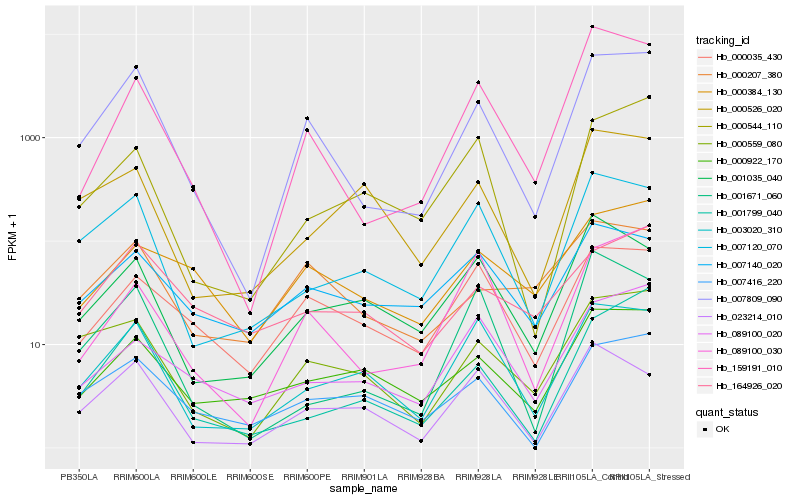
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007809_090 |
0.0 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 2 |
Hb_000559_080 |
0.1568350371 |
- |
- |
PREDICTED: calcineurin B-like protein 4 [Jatropha curcas] |
| 3 |
Hb_159191_010 |
0.1636453266 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 4 |
Hb_007120_070 |
0.1777639041 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 5 |
Hb_003020_310 |
0.1829686358 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_007416_220 |
0.1841447987 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 7 |
Hb_001799_040 |
0.1877539202 |
- |
- |
- |
| 8 |
Hb_000922_170 |
0.195800542 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 9 |
Hb_000035_430 |
0.1962196187 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2-like [Jatropha curcas] |
| 10 |
Hb_089100_030 |
0.196563333 |
- |
- |
- |
| 11 |
Hb_000207_380 |
0.2011813705 |
- |
- |
sucrose transporter 6 [Hevea brasiliensis] |
| 12 |
Hb_000544_110 |
0.2041484639 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 13 |
Hb_023214_010 |
0.2047451224 |
- |
- |
aberrant lateral root formation 1 family protein [Populus trichocarpa] |
| 14 |
Hb_001671_060 |
0.2095326385 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 15 |
Hb_000384_130 |
0.2104550129 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 16 |
Hb_164926_020 |
0.2120237806 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 17 |
Hb_007140_020 |
0.2163065062 |
- |
- |
hypothetical protein CICLE_v10008419mg [Citrus clementina] |
| 18 |
Hb_001035_040 |
0.2165965187 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_089100_020 |
0.2176800848 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 20 |
Hb_000526_020 |
0.2210834185 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |