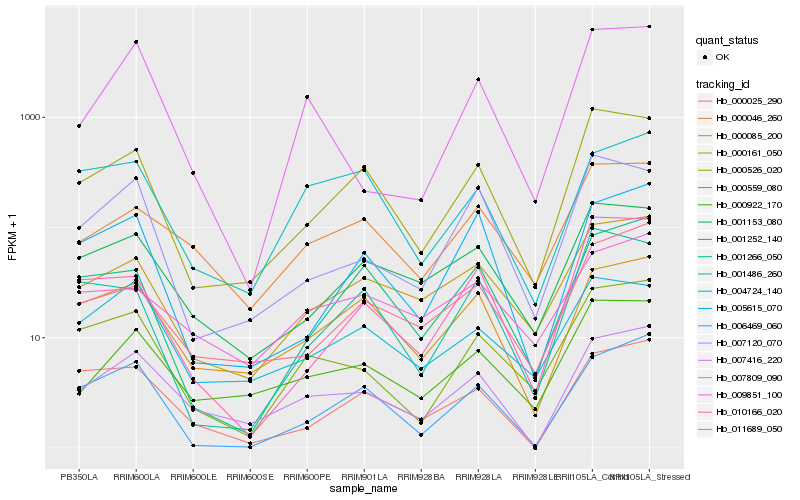
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000559_080 |
0.0 |
- |
- |
PREDICTED: calcineurin B-like protein 4 [Jatropha curcas] |
| 2 |
Hb_000526_020 |
0.1536254 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 3 |
Hb_007809_090 |
0.1568350371 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 4 |
Hb_000161_050 |
0.1575565005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007416_220 |
0.1627239095 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 6 |
Hb_006469_060 |
0.1645194994 |
- |
- |
hypothetical protein POPTR_0005s16470g [Populus trichocarpa] |
| 7 |
Hb_007120_070 |
0.1658083331 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 8 |
Hb_010166_020 |
0.1663205414 |
transcription factor |
TF Family: NF-YC |
ccaat-binding transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000046_260 |
0.1722837582 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 10 |
Hb_001153_080 |
0.1731838441 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL19 [Jatropha curcas] |
| 11 |
Hb_001486_260 |
0.1753399806 |
- |
- |
PREDICTED: uncharacterized protein LOC105632604 [Jatropha curcas] |
| 12 |
Hb_000025_290 |
0.1776457604 |
- |
- |
Uncharacterized protein TCM_031320 [Theobroma cacao] |
| 13 |
Hb_001266_050 |
0.1784555155 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like isoform X2 [Elaeis guineensis] |
| 14 |
Hb_004724_140 |
0.1785216392 |
transcription factor |
TF Family: NF-YB |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 15 |
Hb_001252_140 |
0.1785498069 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_000085_200 |
0.1786264209 |
- |
- |
ATP synthase subunit H protein [Hevea brasiliensis] |
| 17 |
Hb_005615_070 |
0.1804056398 |
- |
- |
Rab7 [Hevea brasiliensis] |
| 18 |
Hb_000922_170 |
0.1807499435 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 19 |
Hb_011689_050 |
0.1827168612 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 20 |
Hb_009851_100 |
0.1839205693 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |