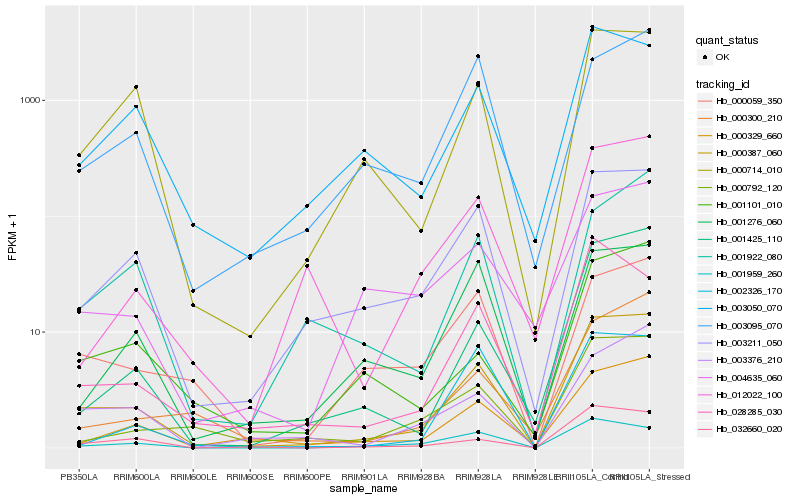
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000387_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_003211_050 |
0.1388022468 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 3 |
Hb_032660_020 |
0.140305122 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 4 |
Hb_000329_660 |
0.1491333295 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000714_010 |
0.1562132376 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 6 |
Hb_001425_110 |
0.1609803612 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |
| 7 |
Hb_000792_120 |
0.164560541 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003376_210 |
0.1659743811 |
- |
- |
- |
| 9 |
Hb_001101_010 |
0.1770161295 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 10 |
Hb_001959_260 |
0.1772917137 |
- |
- |
- |
| 11 |
Hb_003050_070 |
0.1775816466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_012022_100 |
0.1793642979 |
- |
- |
CPRD49 family protein [Populus trichocarpa] |
| 13 |
Hb_000300_210 |
0.1815775502 |
- |
- |
gd2b, putative [Ricinus communis] |
| 14 |
Hb_004635_060 |
0.1822967035 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |
| 15 |
Hb_028285_030 |
0.1828100205 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 16 |
Hb_000059_350 |
0.183251422 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001922_080 |
0.1842059224 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 18 |
Hb_001276_060 |
0.1853585305 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 19 |
Hb_002326_170 |
0.1902760542 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 20 |
Hb_003095_070 |
0.1928368944 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |