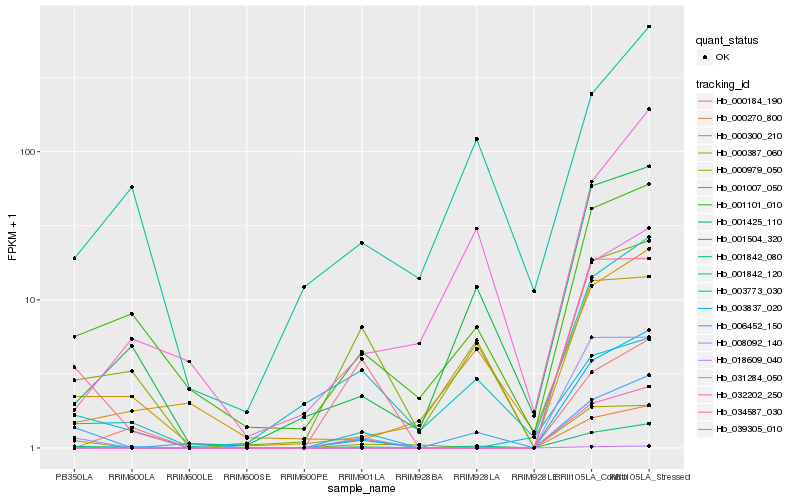
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001504_320 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 2 |
Hb_001425_110 |
0.1485985472 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |
| 3 |
Hb_001842_120 |
0.1658073621 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 4 |
Hb_001101_010 |
0.1759795113 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 5 |
Hb_008092_140 |
0.1905610891 |
- |
- |
PREDICTED: uncharacterized protein LOC103636948 [Zea mays] |
| 6 |
Hb_003837_020 |
0.1921596954 |
- |
- |
PREDICTED: uncharacterized protein LOC103710465 isoform X1 [Phoenix dactylifera] |
| 7 |
Hb_003773_030 |
0.2038436691 |
- |
- |
- |
| 8 |
Hb_001007_050 |
0.2080646043 |
- |
- |
hypothetical protein MIMGU_mgv1a0196712mg, partial [Erythranthe guttata] |
| 9 |
Hb_006452_150 |
0.2081296366 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor 7-like [Jatropha curcas] |
| 10 |
Hb_039305_010 |
0.208475055 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 11 |
Hb_000184_190 |
0.2121020086 |
- |
- |
hypothetical protein VITISV_016970 [Vitis vinifera] |
| 12 |
Hb_000300_210 |
0.2156235639 |
- |
- |
gd2b, putative [Ricinus communis] |
| 13 |
Hb_000979_050 |
0.2185464022 |
- |
- |
- |
| 14 |
Hb_032202_250 |
0.2211298614 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 15 |
Hb_000387_060 |
0.2214632883 |
- |
- |
- |
| 16 |
Hb_001842_080 |
0.2232035506 |
- |
- |
- |
| 17 |
Hb_031284_050 |
0.2242775261 |
- |
- |
- |
| 18 |
Hb_034587_030 |
0.2244256971 |
- |
- |
- |
| 19 |
Hb_018609_040 |
0.2246255488 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 20 |
Hb_000270_800 |
0.2246333838 |
- |
- |
- |