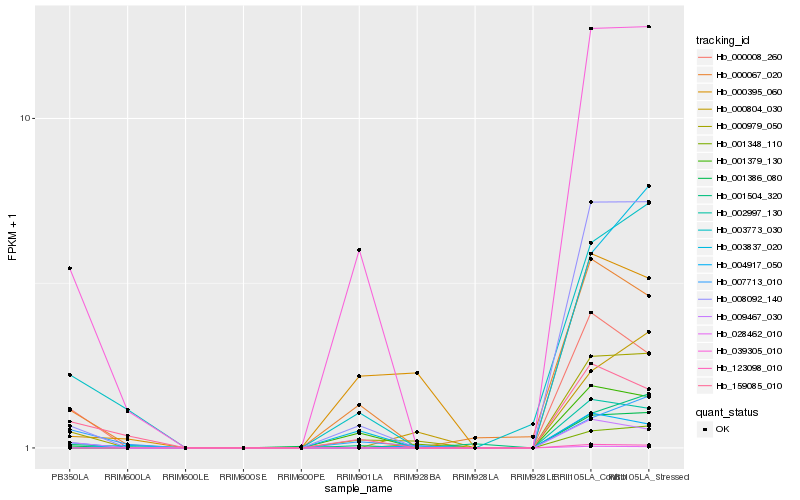
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000979_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_008092_140 |
0.1485888387 |
- |
- |
PREDICTED: uncharacterized protein LOC103636948 [Zea mays] |
| 3 |
Hb_039305_010 |
0.1516741061 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 4 |
Hb_004917_050 |
0.1532054085 |
- |
- |
- |
| 5 |
Hb_009467_030 |
0.1993266538 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 6 |
Hb_000067_020 |
0.2014650042 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 7 |
Hb_001386_080 |
0.2035846361 |
- |
- |
hypothetical protein JCGZ_06052 [Jatropha curcas] |
| 8 |
Hb_003773_030 |
0.2080449223 |
- |
- |
- |
| 9 |
Hb_000008_260 |
0.2118470444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001504_320 |
0.2185464022 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 11 |
Hb_007713_010 |
0.2225384849 |
- |
- |
hypothetical protein JCGZ_18011 [Jatropha curcas] |
| 12 |
Hb_000395_060 |
0.2376884409 |
- |
- |
- |
| 13 |
Hb_003837_020 |
0.2394319325 |
- |
- |
PREDICTED: uncharacterized protein LOC103710465 isoform X1 [Phoenix dactylifera] |
| 14 |
Hb_001348_110 |
0.2403083116 |
- |
- |
PREDICTED: esterase-like [Jatropha curcas] |
| 15 |
Hb_159085_010 |
0.2431547753 |
- |
- |
- |
| 16 |
Hb_000804_030 |
0.2437924866 |
- |
- |
Protein C14orf111, putative [Ricinus communis] |
| 17 |
Hb_001379_130 |
0.2456203873 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 18 |
Hb_002997_130 |
0.2460852795 |
- |
- |
- |
| 19 |
Hb_028462_010 |
0.2461693611 |
- |
- |
PREDICTED: uncharacterized protein LOC101509514 [Cicer arietinum] |
| 20 |
Hb_123098_010 |
0.2461699879 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |