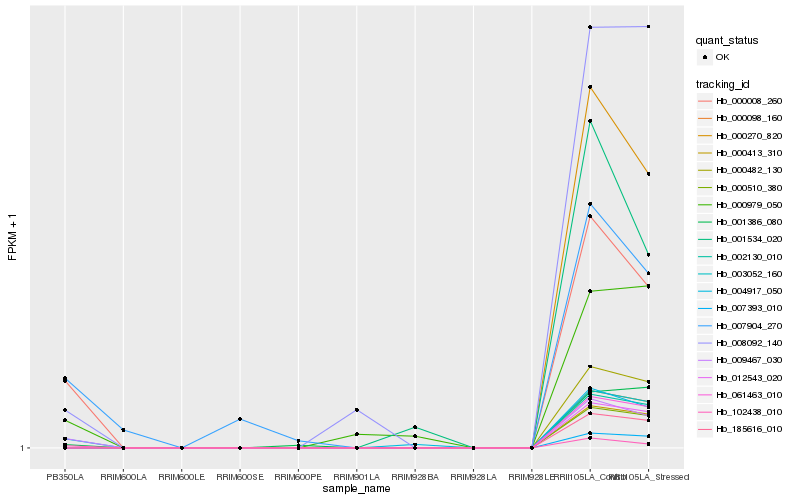
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004917_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_009467_030 |
0.124378962 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 3 |
Hb_000008_260 |
0.1322857471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000979_050 |
0.1532054085 |
- |
- |
- |
| 5 |
Hb_102438_010 |
0.1745488154 |
- |
- |
- |
| 6 |
Hb_001386_080 |
0.2116338774 |
- |
- |
hypothetical protein JCGZ_06052 [Jatropha curcas] |
| 7 |
Hb_008092_140 |
0.2120516705 |
- |
- |
PREDICTED: uncharacterized protein LOC103636948 [Zea mays] |
| 8 |
Hb_001534_020 |
0.2132744794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007904_270 |
0.2272458332 |
- |
- |
- |
| 10 |
Hb_000270_820 |
0.2312237262 |
- |
- |
- |
| 11 |
Hb_000510_380 |
0.233744047 |
- |
- |
- |
| 12 |
Hb_000482_130 |
0.2337447963 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 13 |
Hb_012543_020 |
0.2337454642 |
- |
- |
- |
| 14 |
Hb_185616_010 |
0.2337465356 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 15 |
Hb_061463_010 |
0.2337522527 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 16 |
Hb_002130_010 |
0.2337542292 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 17 |
Hb_000098_160 |
0.2337641281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003052_160 |
0.2337641281 |
- |
- |
- |
| 19 |
Hb_000413_310 |
0.2337660824 |
- |
- |
- |
| 20 |
Hb_007393_010 |
0.2337811807 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |