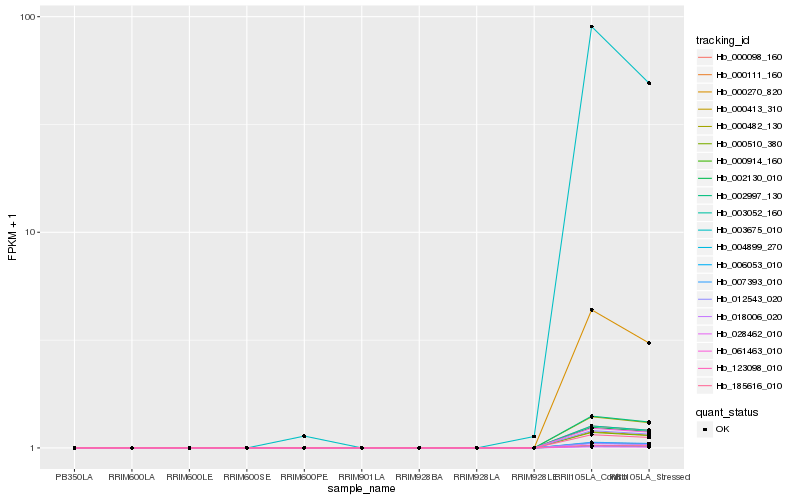
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002997_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_028462_010 |
0.0003642905 |
- |
- |
PREDICTED: uncharacterized protein LOC101509514 [Cicer arietinum] |
| 3 |
Hb_123098_010 |
0.0003669985 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_004899_270 |
0.0003875599 |
- |
- |
PREDICTED: beta-galactosidase 15-like [Cucumis sativus] |
| 5 |
Hb_000111_160 |
0.0004154329 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 6 |
Hb_018006_020 |
0.0004768542 |
- |
- |
PREDICTED: uncharacterized protein LOC105641417 [Jatropha curcas] |
| 7 |
Hb_006053_010 |
0.0004940828 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 8 |
Hb_000914_160 |
0.0005063856 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Glycine max] |
| 9 |
Hb_007393_010 |
0.0005253368 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 10 |
Hb_000413_310 |
0.0006234418 |
- |
- |
- |
| 11 |
Hb_000098_160 |
0.0006361585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003052_160 |
0.0006361585 |
- |
- |
- |
| 13 |
Hb_002130_010 |
0.0007006343 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 14 |
Hb_061463_010 |
0.0007135206 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 15 |
Hb_185616_010 |
0.0007508194 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 16 |
Hb_012543_020 |
0.0007578135 |
- |
- |
- |
| 17 |
Hb_000482_130 |
0.0007621738 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 18 |
Hb_000510_380 |
0.0007670663 |
- |
- |
- |
| 19 |
Hb_000270_820 |
0.0526038109 |
- |
- |
- |
| 20 |
Hb_003675_010 |
0.0839575925 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |