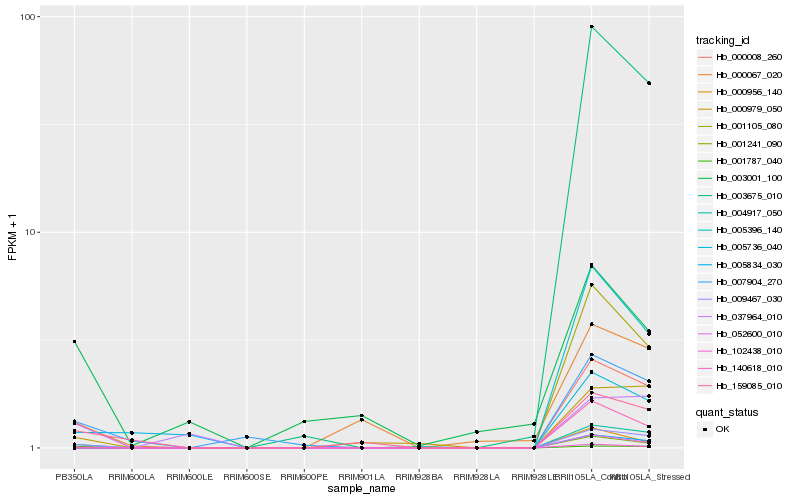
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_102438_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000008_260 |
0.0819698869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009467_030 |
0.1022514247 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 4 |
Hb_004917_050 |
0.1745488154 |
- |
- |
- |
| 5 |
Hb_007904_270 |
0.2086186569 |
- |
- |
- |
| 6 |
Hb_001787_040 |
0.2209548894 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_159085_010 |
0.2320042267 |
- |
- |
- |
| 8 |
Hb_000067_020 |
0.268124516 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 9 |
Hb_140618_010 |
0.2681363089 |
- |
- |
- |
| 10 |
Hb_001241_090 |
0.2681363113 |
- |
- |
PREDICTED: uncharacterized protein LOC105775709 [Gossypium raimondii] |
| 11 |
Hb_052600_010 |
0.2681363522 |
- |
- |
- |
| 12 |
Hb_005396_140 |
0.2681470602 |
- |
- |
- |
| 13 |
Hb_001105_080 |
0.2682546192 |
- |
- |
- |
| 14 |
Hb_000979_050 |
0.2697612741 |
- |
- |
- |
| 15 |
Hb_003001_100 |
0.2703239722 |
- |
- |
PREDICTED: uncharacterized protein LOC103334281 [Prunus mume] |
| 16 |
Hb_005834_030 |
0.2735835403 |
- |
- |
- |
| 17 |
Hb_003675_010 |
0.2764163106 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_000956_140 |
0.2774306951 |
- |
- |
- |
| 19 |
Hb_005736_040 |
0.2791338844 |
- |
- |
- |
| 20 |
Hb_037964_010 |
0.2798043412 |
- |
- |
hypothetical protein MIMGU_mgv1a000906mg [Erythranthe guttata] |