| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
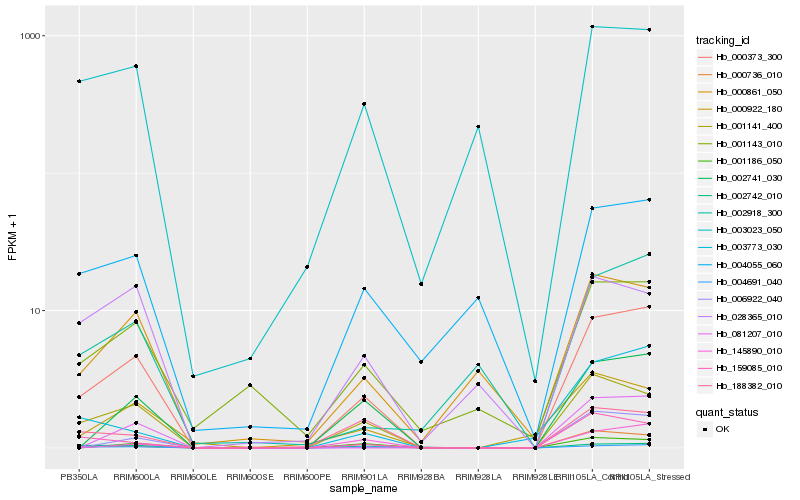
Hb_000373_300 |
0.0 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 2 |
Hb_001141_400 |
0.1539355832 |
- |
- |
hypothetical protein B456_007G2854002, partial [Gossypium raimondii] |
| 3 |
Hb_002742_010 |
0.1891689237 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 4 |
Hb_002741_030 |
0.1948927724 |
- |
- |
- |
| 5 |
Hb_000861_050 |
0.2078497866 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 6 |
Hb_001143_010 |
0.2116614172 |
- |
- |
- |
| 7 |
Hb_159085_010 |
0.2116994288 |
- |
- |
- |
| 8 |
Hb_003773_030 |
0.217877435 |
- |
- |
- |
| 9 |
Hb_000922_180 |
0.2212175435 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 10 |
Hb_001186_050 |
0.2236898393 |
- |
- |
- |
| 11 |
Hb_004055_060 |
0.2239794958 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 12 |
Hb_002918_300 |
0.2245390634 |
- |
- |
- |
| 13 |
Hb_004691_040 |
0.2250420686 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 14 |
Hb_003023_050 |
0.2263427854 |
- |
- |
hypothetical protein POPTR_0013s10320g [Populus trichocarpa] |
| 15 |
Hb_000736_010 |
0.2286933298 |
- |
- |
PREDICTED: uncharacterized protein LOC105762083 [Gossypium raimondii] |
| 16 |
Hb_145890_010 |
0.2411636789 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_188382_010 |
0.2434943387 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 18 |
Hb_028365_010 |
0.2440037561 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 19 |
Hb_081207_010 |
0.2455084673 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 20 |
Hb_006922_040 |
0.2464567436 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |