| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
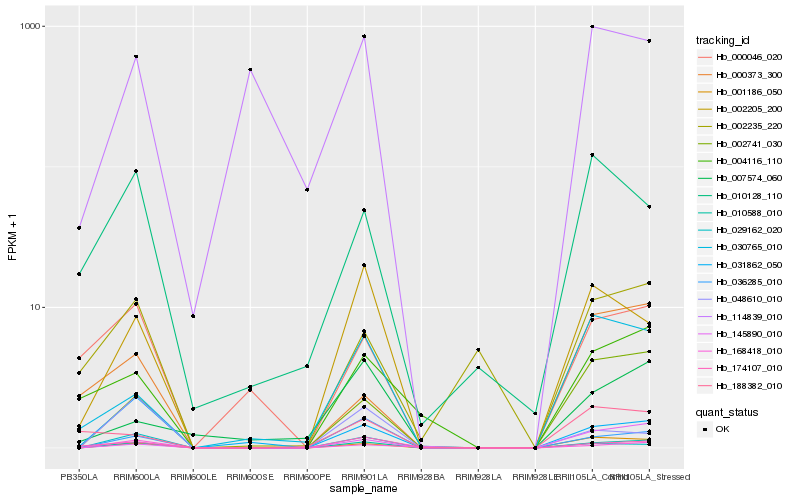
Hb_010588_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001186_050 |
0.1800001948 |
- |
- |
- |
| 3 |
Hb_002741_030 |
0.1848882156 |
- |
- |
- |
| 4 |
Hb_036285_010 |
0.1981407699 |
- |
- |
- |
| 5 |
Hb_168418_010 |
0.2277921257 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 6 |
Hb_002205_200 |
0.2308459274 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 7 |
Hb_174107_010 |
0.2454440881 |
- |
- |
PREDICTED: uncharacterized protein LOC105775323 [Gossypium raimondii] |
| 8 |
Hb_145890_010 |
0.247913924 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_004116_110 |
0.2533040744 |
- |
- |
hypothetical protein MTR_3g064100 [Medicago truncatula] |
| 10 |
Hb_030765_010 |
0.2568034769 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter G family member 11-like isoform X1 [Solanum lycopersicum] |
| 11 |
Hb_031862_050 |
0.2661703895 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 12 |
Hb_000046_020 |
0.2865532426 |
- |
- |
- |
| 13 |
Hb_029162_020 |
0.2874017398 |
- |
- |
- |
| 14 |
Hb_007574_060 |
0.2896583998 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 27-like [Jatropha curcas] |
| 15 |
Hb_002235_220 |
0.2934264181 |
- |
- |
PREDICTED: pectinesterase 31-like [Jatropha curcas] |
| 16 |
Hb_010128_110 |
0.2954080233 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 17 |
Hb_000373_300 |
0.3021084963 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 18 |
Hb_048610_010 |
0.3045300246 |
- |
- |
- |
| 19 |
Hb_114839_010 |
0.3085690501 |
- |
- |
- |
| 20 |
Hb_188382_010 |
0.3086811342 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |