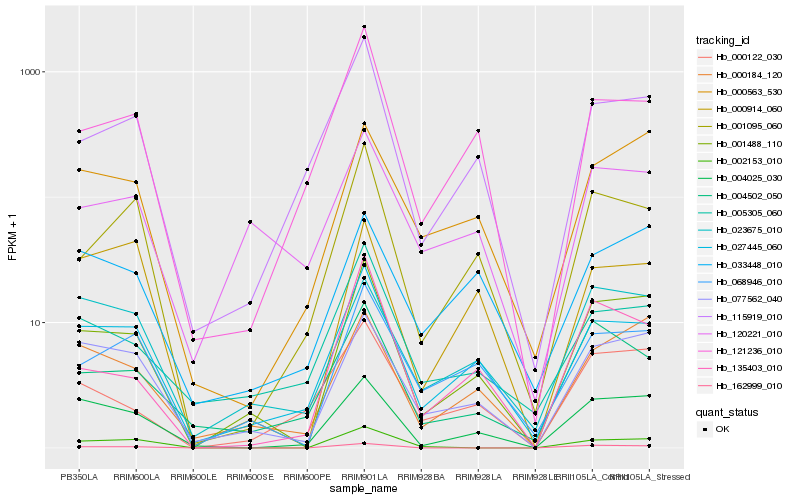
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_077562_040 |
0.1373349355 |
- |
- |
hypothetical protein JCGZ_24573 [Jatropha curcas] |
| 3 |
Hb_027445_060 |
0.1450691616 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_000184_120 |
0.14542429 |
- |
- |
hypothetical protein JCGZ_18526 [Jatropha curcas] |
| 5 |
Hb_068946_010 |
0.1494888857 |
- |
- |
hypothetical protein CICLE_v100122621mg, partial [Citrus clementina] |
| 6 |
Hb_004025_030 |
0.1523069078 |
- |
- |
- |
| 7 |
Hb_000563_530 |
0.1691340151 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 8 |
Hb_001095_060 |
0.1718408049 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 9 |
Hb_115919_010 |
0.1767737984 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_135403_010 |
0.1772569699 |
- |
- |
HEV1.1 [Hevea brasiliensis] |
| 11 |
Hb_000122_030 |
0.1779946387 |
- |
- |
PREDICTED: uncharacterized protein LOC104606081, partial [Nelumbo nucifera] |
| 12 |
Hb_162999_010 |
0.1782110476 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 13 |
Hb_023675_010 |
0.1812279699 |
- |
- |
- |
| 14 |
Hb_002153_010 |
0.1837496684 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 15 |
Hb_004502_050 |
0.1858947764 |
- |
- |
hypothetical protein POPTR_1488s00200g [Populus trichocarpa] |
| 16 |
Hb_121236_010 |
0.1867239744 |
- |
- |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 17 |
Hb_033448_010 |
0.1923471617 |
- |
- |
hypothetical protein JCGZ_19772 [Jatropha curcas] |
| 18 |
Hb_005305_060 |
0.1941038111 |
- |
- |
hypothetical protein MIMGU_mgv1a015111mg [Erythranthe guttata] |
| 19 |
Hb_120221_010 |
0.1971390803 |
- |
- |
- |
| 20 |
Hb_000914_060 |
0.1987087034 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |