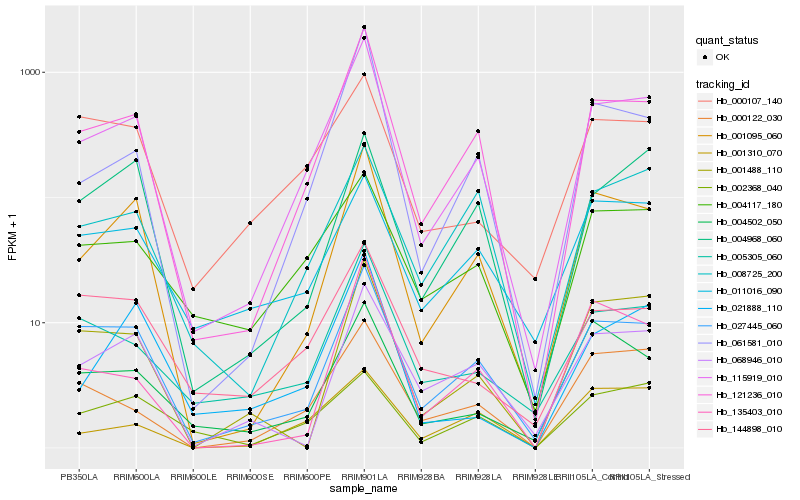
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115919_010 |
0.0 |
- |
- |
unknown [Lotus japonicus] |
| 2 |
Hb_121236_010 |
0.0667305932 |
- |
- |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_027445_060 |
0.1157118981 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_001095_060 |
0.1205172126 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 5 |
Hb_061581_010 |
0.1421324301 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_005305_060 |
0.1549066243 |
- |
- |
hypothetical protein MIMGU_mgv1a015111mg [Erythranthe guttata] |
| 7 |
Hb_000122_030 |
0.1602966522 |
- |
- |
PREDICTED: uncharacterized protein LOC104606081, partial [Nelumbo nucifera] |
| 8 |
Hb_004117_180 |
0.1611902076 |
- |
- |
- |
| 9 |
Hb_008725_200 |
0.1734598495 |
- |
- |
PREDICTED: uncharacterized protein LOC105642590 [Jatropha curcas] |
| 10 |
Hb_001488_110 |
0.1767737984 |
- |
- |
- |
| 11 |
Hb_004502_050 |
0.1773861354 |
- |
- |
hypothetical protein POPTR_1488s00200g [Populus trichocarpa] |
| 12 |
Hb_068946_010 |
0.1801694437 |
- |
- |
hypothetical protein CICLE_v100122621mg, partial [Citrus clementina] |
| 13 |
Hb_144898_010 |
0.1818324348 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 14 |
Hb_021888_110 |
0.1823843049 |
- |
- |
- |
| 15 |
Hb_001310_070 |
0.1853678185 |
- |
- |
PREDICTED: uncharacterized protein LOC105639838 [Jatropha curcas] |
| 16 |
Hb_135403_010 |
0.1860278451 |
- |
- |
HEV1.1 [Hevea brasiliensis] |
| 17 |
Hb_000107_140 |
0.1862404211 |
- |
- |
- |
| 18 |
Hb_002368_040 |
0.1862456128 |
- |
- |
hypothetical protein PRUPE_ppa019172mg [Prunus persica] |
| 19 |
Hb_011016_090 |
0.1872932157 |
- |
- |
PREDICTED: 50S ribosomal protein L24, chloroplastic [Populus euphratica] |
| 20 |
Hb_004968_060 |
0.1879203754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |