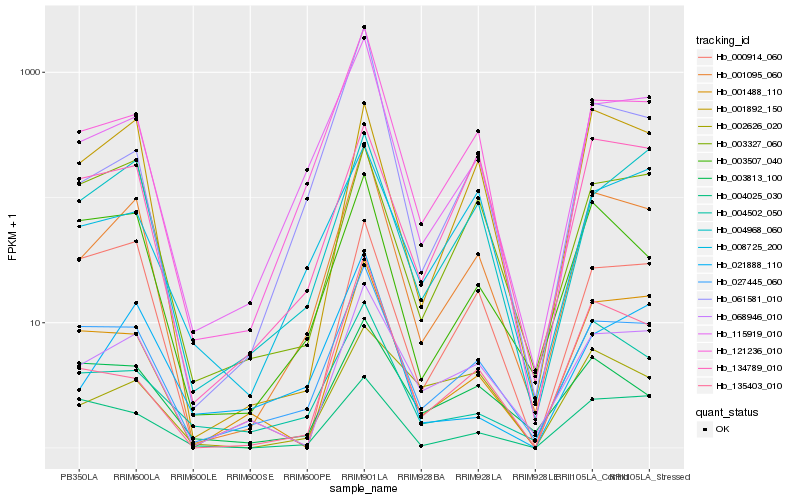
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001095_060 |
0.0 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 2 |
Hb_121236_010 |
0.1202899694 |
- |
- |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_115919_010 |
0.1205172126 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_027445_060 |
0.1362152085 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_068946_010 |
0.1451316679 |
- |
- |
hypothetical protein CICLE_v100122621mg, partial [Citrus clementina] |
| 6 |
Hb_061581_010 |
0.1717492364 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_001488_110 |
0.1718408049 |
- |
- |
- |
| 8 |
Hb_001892_150 |
0.1747094702 |
- |
- |
- |
| 9 |
Hb_003507_040 |
0.1748174628 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 10 |
Hb_135403_010 |
0.1809469432 |
- |
- |
HEV1.1 [Hevea brasiliensis] |
| 11 |
Hb_004502_050 |
0.1814607489 |
- |
- |
hypothetical protein POPTR_1488s00200g [Populus trichocarpa] |
| 12 |
Hb_004968_060 |
0.1854402303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000914_060 |
0.1995314817 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 14 |
Hb_003327_060 |
0.2019142921 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 15 |
Hb_021888_110 |
0.2037396832 |
- |
- |
- |
| 16 |
Hb_134789_010 |
0.2043142293 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 17 |
Hb_003813_100 |
0.2049375344 |
- |
- |
PREDICTED: proteasome subunit beta type-3-A [Cucumis sativus] |
| 18 |
Hb_008725_200 |
0.2079147595 |
- |
- |
PREDICTED: uncharacterized protein LOC105642590 [Jatropha curcas] |
| 19 |
Hb_004025_030 |
0.2099674478 |
- |
- |
- |
| 20 |
Hb_002626_020 |
0.2131315081 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |