| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
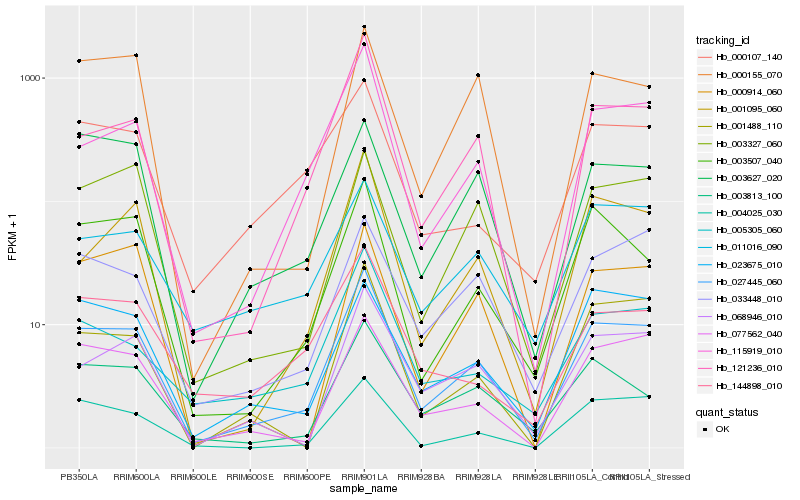
Hb_027445_060 |
0.0 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_121236_010 |
0.1112983686 |
- |
- |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_115919_010 |
0.1157118981 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_068946_010 |
0.1307265212 |
- |
- |
hypothetical protein CICLE_v100122621mg, partial [Citrus clementina] |
| 5 |
Hb_001095_060 |
0.1362152085 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 6 |
Hb_003813_100 |
0.1417769505 |
- |
- |
PREDICTED: proteasome subunit beta type-3-A [Cucumis sativus] |
| 7 |
Hb_005305_060 |
0.1426439745 |
- |
- |
hypothetical protein MIMGU_mgv1a015111mg [Erythranthe guttata] |
| 8 |
Hb_003507_040 |
0.143777108 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 9 |
Hb_001488_110 |
0.1450691616 |
- |
- |
- |
| 10 |
Hb_077562_040 |
0.1453415505 |
- |
- |
hypothetical protein JCGZ_24573 [Jatropha curcas] |
| 11 |
Hb_004025_030 |
0.1508897143 |
- |
- |
- |
| 12 |
Hb_144898_010 |
0.1546990133 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 13 |
Hb_033448_010 |
0.1547052161 |
- |
- |
hypothetical protein JCGZ_19772 [Jatropha curcas] |
| 14 |
Hb_000155_070 |
0.1587055421 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 15 |
Hb_000107_140 |
0.1592716613 |
- |
- |
- |
| 16 |
Hb_011016_090 |
0.1597377028 |
- |
- |
PREDICTED: 50S ribosomal protein L24, chloroplastic [Populus euphratica] |
| 17 |
Hb_000914_060 |
0.1599538185 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 18 |
Hb_003327_060 |
0.1616207815 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 19 |
Hb_023675_010 |
0.1626162538 |
- |
- |
- |
| 20 |
Hb_003627_020 |
0.162999944 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |