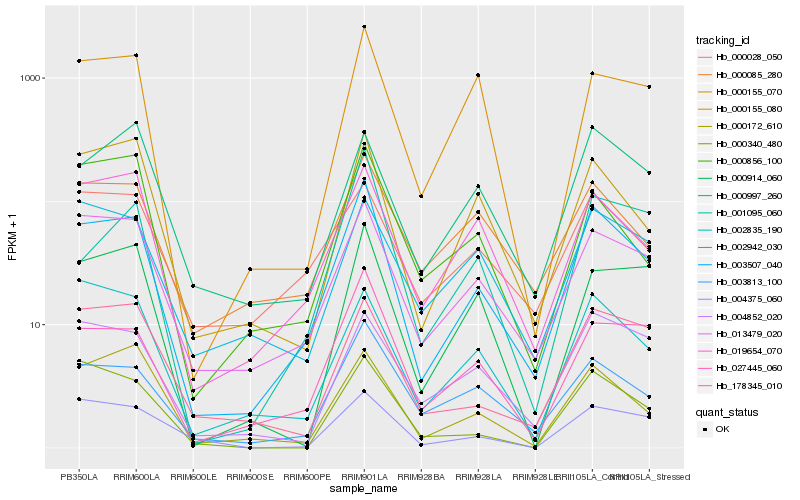
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003507_040 |
0.0 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 2 |
Hb_013479_020 |
0.1317391334 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 3 |
Hb_003813_100 |
0.1354592119 |
- |
- |
PREDICTED: proteasome subunit beta type-3-A [Cucumis sativus] |
| 4 |
Hb_027445_060 |
0.143777108 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_000155_080 |
0.1490598148 |
- |
- |
guanylate-binding family protein [Populus trichocarpa] |
| 6 |
Hb_019654_070 |
0.1493661892 |
- |
- |
PREDICTED: probable alpha-amylase 2 [Jatropha curcas] |
| 7 |
Hb_000172_610 |
0.1593059403 |
- |
- |
hypothetical protein POPTR_0017s02240g, partial [Populus trichocarpa] |
| 8 |
Hb_002835_190 |
0.1663741345 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 9 |
Hb_004375_060 |
0.1697785733 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_178345_010 |
0.1700023555 |
- |
- |
hypothetical protein PHAVU_004G084200g [Phaseolus vulgaris] |
| 11 |
Hb_000085_280 |
0.1703113923 |
- |
- |
PREDICTED: beta-(1,2)-xylosyltransferase [Jatropha curcas] |
| 12 |
Hb_004852_020 |
0.1707986864 |
- |
- |
- |
| 13 |
Hb_000155_070 |
0.1721852912 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 14 |
Hb_000856_100 |
0.1723910364 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 15 |
Hb_001095_060 |
0.1748174628 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 16 |
Hb_000997_260 |
0.1748249053 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000340_480 |
0.1752795603 |
- |
- |
- |
| 18 |
Hb_000914_060 |
0.1764596091 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 19 |
Hb_002942_030 |
0.177014175 |
- |
- |
hypothetical protein EUGRSUZ_K000651 [Eucalyptus grandis] |
| 20 |
Hb_000028_050 |
0.1772324865 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |