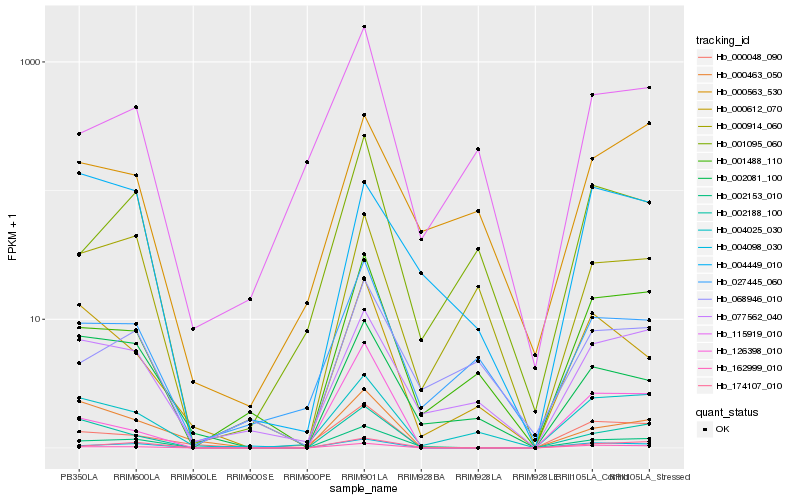
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002153_010 |
0.0 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 2 |
Hb_174107_010 |
0.1221682008 |
- |
- |
PREDICTED: uncharacterized protein LOC105775323 [Gossypium raimondii] |
| 3 |
Hb_162999_010 |
0.1541436538 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 4 |
Hb_001488_110 |
0.1837496684 |
- |
- |
- |
| 5 |
Hb_002188_100 |
0.1850545904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_077562_040 |
0.1950206895 |
- |
- |
hypothetical protein JCGZ_24573 [Jatropha curcas] |
| 7 |
Hb_000048_090 |
0.2030337874 |
transcription factor |
TF Family: SET |
hypothetical protein JCGZ_06559 [Jatropha curcas] |
| 8 |
Hb_004025_030 |
0.2073915415 |
- |
- |
- |
| 9 |
Hb_027445_060 |
0.212827544 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_002081_100 |
0.22120718 |
- |
- |
- |
| 11 |
Hb_001095_060 |
0.2215403619 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 12 |
Hb_126398_010 |
0.2234365971 |
- |
- |
- |
| 13 |
Hb_068946_010 |
0.2284837626 |
- |
- |
hypothetical protein CICLE_v100122621mg, partial [Citrus clementina] |
| 14 |
Hb_000463_050 |
0.2285334384 |
- |
- |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 15 |
Hb_004098_030 |
0.2308243389 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 16 |
Hb_000563_530 |
0.2319619012 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 17 |
Hb_000612_070 |
0.2331538435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000914_060 |
0.2368356628 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 19 |
Hb_004449_010 |
0.2370780505 |
- |
- |
lipase [Ricinus communis] |
| 20 |
Hb_115919_010 |
0.2387765697 |
- |
- |
unknown [Lotus japonicus] |