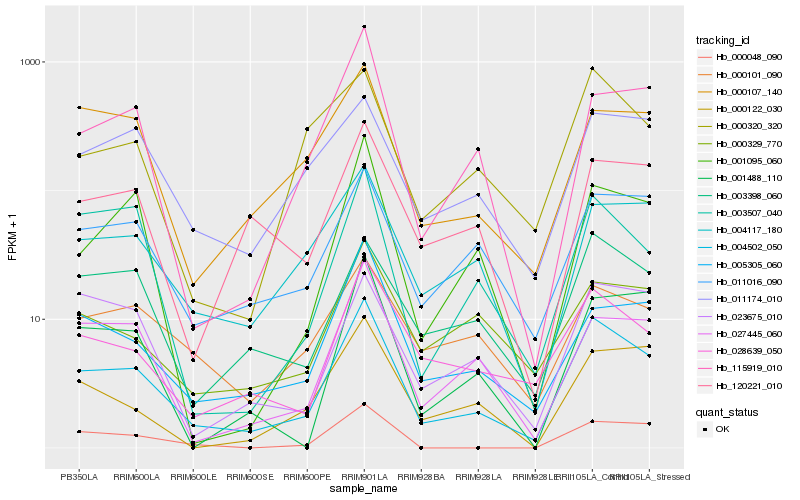
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004502_050 |
0.0 |
- |
- |
hypothetical protein POPTR_1488s00200g [Populus trichocarpa] |
| 2 |
Hb_028639_050 |
0.1555287533 |
- |
- |
Anopheles gambiae str. PEST AGAP012660-PA [Anopheles gambiae str. PEST] |
| 3 |
Hb_011016_090 |
0.1660579813 |
- |
- |
PREDICTED: 50S ribosomal protein L24, chloroplastic [Populus euphratica] |
| 4 |
Hb_005305_060 |
0.1667156675 |
- |
- |
hypothetical protein MIMGU_mgv1a015111mg [Erythranthe guttata] |
| 5 |
Hb_003398_060 |
0.1670310171 |
- |
- |
PREDICTED: rRNA methyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_027445_060 |
0.1699853414 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_004117_180 |
0.1707617124 |
- |
- |
- |
| 8 |
Hb_115919_010 |
0.1773861354 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_003507_040 |
0.1779075245 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 10 |
Hb_001095_060 |
0.1814607489 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 11 |
Hb_000107_140 |
0.1829418827 |
- |
- |
- |
| 12 |
Hb_120221_010 |
0.184161043 |
- |
- |
- |
| 13 |
Hb_001488_110 |
0.1858947764 |
- |
- |
- |
| 14 |
Hb_000320_320 |
0.1869620464 |
- |
- |
hypothetical protein POPTR_0004s01360g [Populus trichocarpa] |
| 15 |
Hb_000122_030 |
0.187873562 |
- |
- |
PREDICTED: uncharacterized protein LOC104606081, partial [Nelumbo nucifera] |
| 16 |
Hb_011174_010 |
0.1883165691 |
- |
- |
hypothetical protein JCGZ_10122 [Jatropha curcas] |
| 17 |
Hb_023675_010 |
0.1888943243 |
- |
- |
- |
| 18 |
Hb_000101_090 |
0.189039272 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Camelina sativa] |
| 19 |
Hb_000048_090 |
0.1903272708 |
transcription factor |
TF Family: SET |
hypothetical protein JCGZ_06559 [Jatropha curcas] |
| 20 |
Hb_000329_770 |
0.1905419544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |