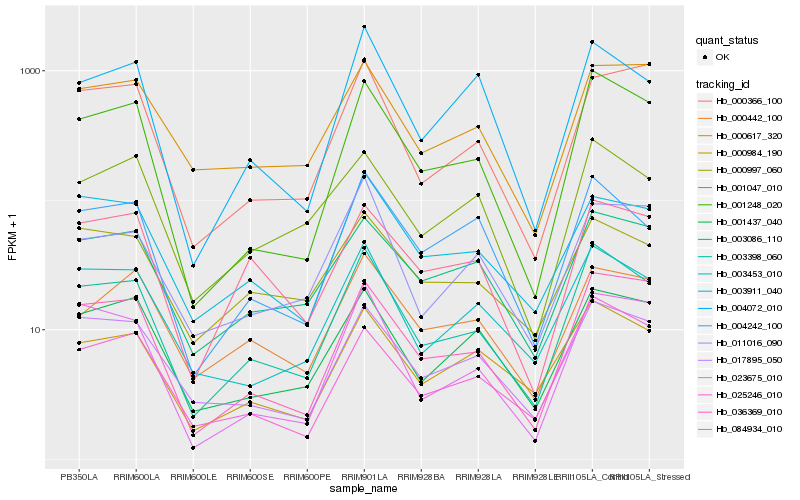
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003398_060 |
0.0 |
- |
- |
PREDICTED: rRNA methyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_001248_020 |
0.081440461 |
- |
- |
PREDICTED: uncharacterized protein At2g23090-like [Jatropha curcas] |
| 3 |
Hb_036369_010 |
0.0984288311 |
- |
- |
- |
| 4 |
Hb_003453_010 |
0.1044729614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000984_190 |
0.1059568453 |
- |
- |
- |
| 6 |
Hb_017895_050 |
0.1083273795 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: UPF0160 protein MYG1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_025246_010 |
0.1115304923 |
- |
- |
- |
| 8 |
Hb_004242_100 |
0.1119425363 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_023675_010 |
0.1163988056 |
- |
- |
- |
| 10 |
Hb_001437_040 |
0.1188108558 |
- |
- |
PREDICTED: ATPase GET3-like [Vitis vinifera] |
| 11 |
Hb_003086_110 |
0.1188167799 |
- |
- |
PREDICTED: dihydroneopterin aldolase-like isoform X2 [Prunus mume] |
| 12 |
Hb_004072_010 |
0.1227307506 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001047_010 |
0.1240184466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003911_040 |
0.1245176063 |
- |
- |
PREDICTED: putative H/ACA ribonucleoprotein complex subunit 1-like protein 1 [Jatropha curcas] |
| 15 |
Hb_084934_010 |
0.1277050427 |
- |
- |
hypothetical protein JCGZ_22225 [Jatropha curcas] |
| 16 |
Hb_011016_090 |
0.1312062234 |
- |
- |
PREDICTED: 50S ribosomal protein L24, chloroplastic [Populus euphratica] |
| 17 |
Hb_000617_320 |
0.1321859182 |
- |
- |
40S ribosomal protein S21B [Hevea brasiliensis] |
| 18 |
Hb_000997_060 |
0.1325180188 |
- |
- |
PREDICTED: DNA repair protein XRCC4 [Jatropha curcas] |
| 19 |
Hb_000366_100 |
0.1338342858 |
- |
- |
60S ribosomal protein L7aA [Hevea brasiliensis] |
| 20 |
Hb_000442_100 |
0.1339357353 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |