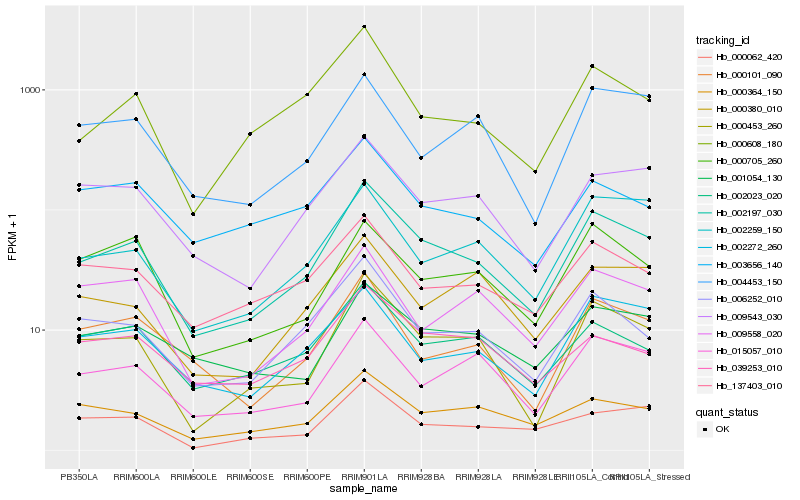
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002197_030 |
0.0 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000453_260 |
0.1000880342 |
- |
- |
PREDICTED: armadillo repeat-containing protein 7 isoform X1 [Vitis vinifera] |
| 3 |
Hb_006252_010 |
0.1100162868 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009543_030 |
0.1218782967 |
- |
- |
prefoldin subunit 1 [Jatropha curcas] |
| 5 |
Hb_002023_020 |
0.124067615 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |
| 6 |
Hb_000608_180 |
0.1260033422 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Tarenaya hassleriana] |
| 7 |
Hb_000364_150 |
0.1267487973 |
- |
- |
- |
| 8 |
Hb_015057_010 |
0.1270222598 |
- |
- |
PREDICTED: uncharacterized exonuclease domain-containing protein At3g15140 [Jatropha curcas] |
| 9 |
Hb_137403_010 |
0.1302886007 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 10 |
Hb_002272_260 |
0.1314155373 |
- |
- |
- |
| 11 |
Hb_000101_090 |
0.1319111862 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Camelina sativa] |
| 12 |
Hb_000062_420 |
0.1345159701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001054_130 |
0.1351309181 |
- |
- |
Protein SSU72, putative [Ricinus communis] |
| 14 |
Hb_039253_010 |
0.1357091204 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002259_150 |
0.1360045634 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 16 |
Hb_000705_260 |
0.1430419547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000380_010 |
0.1439095718 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X3 [Jatropha curcas] |
| 18 |
Hb_009558_020 |
0.1440962807 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PRT6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004453_150 |
0.1455554768 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 20 |
Hb_003656_140 |
0.1470250903 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |