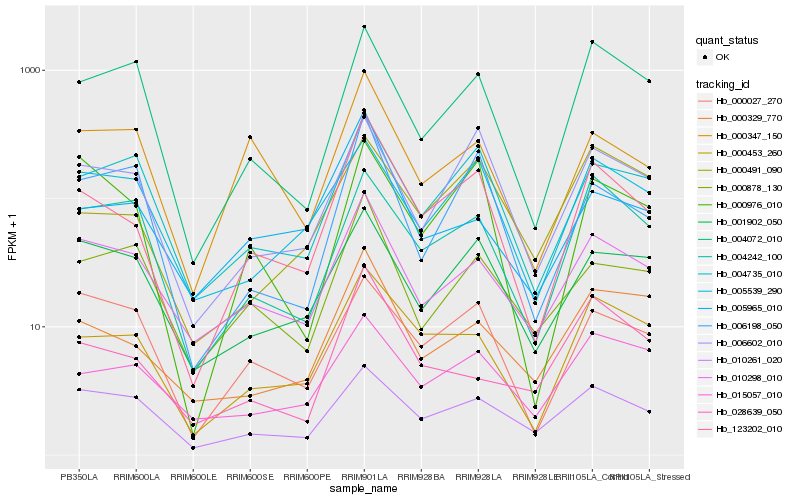
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123202_010 |
0.0 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 2 |
Hb_000453_260 |
0.1300142417 |
- |
- |
PREDICTED: armadillo repeat-containing protein 7 isoform X1 [Vitis vinifera] |
| 3 |
Hb_010298_010 |
0.1622688141 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 [Jatropha curcas] |
| 4 |
Hb_000329_770 |
0.162949294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006602_010 |
0.164621753 |
- |
- |
PREDICTED: tobamovirus multiplication protein 3 [Jatropha curcas] |
| 6 |
Hb_004735_010 |
0.167366132 |
- |
- |
PREDICTED: protein arginine N-methyltransferase PRMT10 [Vitis vinifera] |
| 7 |
Hb_005965_010 |
0.1792160001 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 8 |
Hb_000491_090 |
0.1802125848 |
- |
- |
hypothetical protein B456_008G219400 [Gossypium raimondii] |
| 9 |
Hb_006198_050 |
0.1806957913 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000976_010 |
0.1808003784 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 4 [Jatropha curcas] |
| 11 |
Hb_010261_020 |
0.1814323401 |
- |
- |
hypothetical protein JCGZ_17258 [Jatropha curcas] |
| 12 |
Hb_001902_050 |
0.1836840216 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105648450 [Jatropha curcas] |
| 13 |
Hb_004072_010 |
0.1844571304 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_015057_010 |
0.1848078825 |
- |
- |
PREDICTED: uncharacterized exonuclease domain-containing protein At3g15140 [Jatropha curcas] |
| 15 |
Hb_000347_150 |
0.1849341422 |
- |
- |
Nucleolysin TIAR [Morus notabilis] |
| 16 |
Hb_005539_290 |
0.1874045382 |
- |
- |
unknown [Lotus japonicus] |
| 17 |
Hb_028639_050 |
0.1880208857 |
- |
- |
Anopheles gambiae str. PEST AGAP012660-PA [Anopheles gambiae str. PEST] |
| 18 |
Hb_000878_130 |
0.1889077747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000027_270 |
0.1903121967 |
- |
- |
PREDICTED: zinc finger protein 6 [Jatropha curcas] |
| 20 |
Hb_004242_100 |
0.1907825109 |
- |
- |
RNA binding protein, putative [Ricinus communis] |