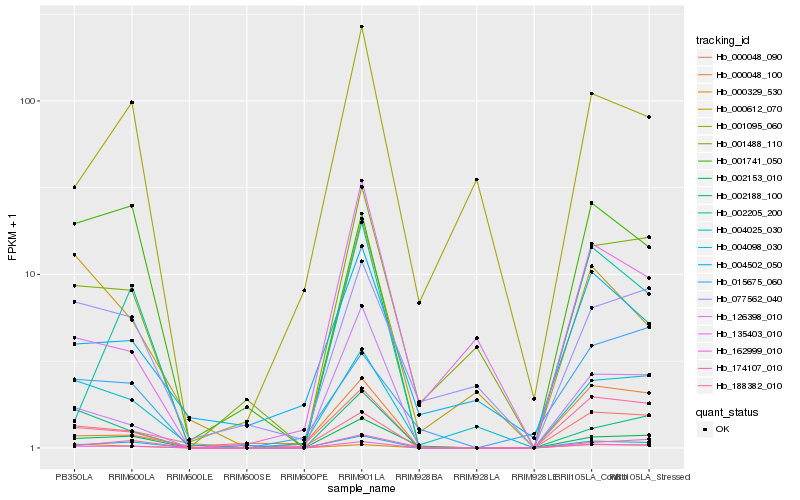
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162999_010 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_000048_090 |
0.1415023113 |
transcription factor |
TF Family: SET |
hypothetical protein JCGZ_06559 [Jatropha curcas] |
| 3 |
Hb_002153_010 |
0.1541436538 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 4 |
Hb_126398_010 |
0.1651268344 |
- |
- |
- |
| 5 |
Hb_001488_110 |
0.1782110476 |
- |
- |
- |
| 6 |
Hb_174107_010 |
0.1896036107 |
- |
- |
PREDICTED: uncharacterized protein LOC105775323 [Gossypium raimondii] |
| 7 |
Hb_002188_100 |
0.1951464997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_188382_010 |
0.1987656952 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 9 |
Hb_004025_030 |
0.2006590095 |
- |
- |
- |
| 10 |
Hb_000329_530 |
0.2023899721 |
- |
- |
Uncharacterized protein TCM_038736 [Theobroma cacao] |
| 11 |
Hb_002205_200 |
0.2026376789 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 12 |
Hb_000048_100 |
0.2057186227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004098_030 |
0.2096877068 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 14 |
Hb_004502_050 |
0.2246556845 |
- |
- |
hypothetical protein POPTR_1488s00200g [Populus trichocarpa] |
| 15 |
Hb_000612_070 |
0.2295743167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_077562_040 |
0.234621303 |
- |
- |
hypothetical protein JCGZ_24573 [Jatropha curcas] |
| 17 |
Hb_001095_060 |
0.2356034103 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 18 |
Hb_001741_050 |
0.237509845 |
- |
- |
- |
| 19 |
Hb_135403_010 |
0.2390113467 |
- |
- |
HEV1.1 [Hevea brasiliensis] |
| 20 |
Hb_015675_060 |
0.2500218467 |
- |
- |
- |