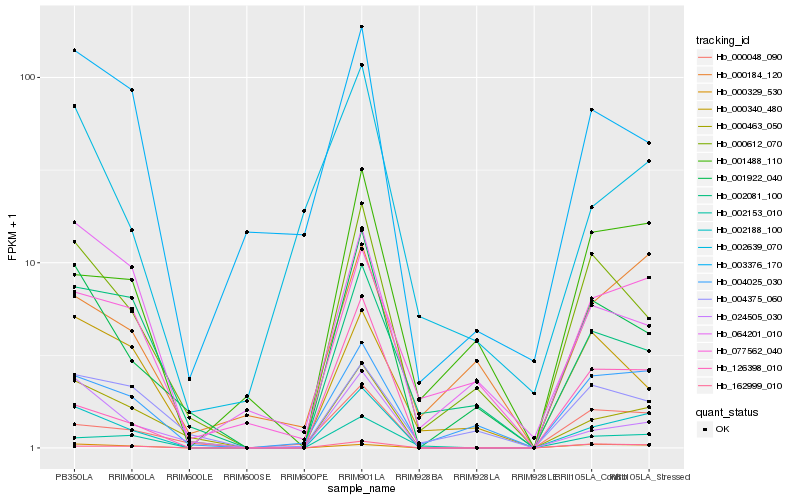
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002188_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000463_050 |
0.1484321503 |
- |
- |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 3 |
Hb_024505_030 |
0.1820132524 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002153_010 |
0.1850545904 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 5 |
Hb_001922_040 |
0.190439476 |
- |
- |
reticulon family protein [Populus trichocarpa] |
| 6 |
Hb_000329_530 |
0.1917429922 |
- |
- |
Uncharacterized protein TCM_038736 [Theobroma cacao] |
| 7 |
Hb_162999_010 |
0.1951464997 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 8 |
Hb_004025_030 |
0.2007323152 |
- |
- |
- |
| 9 |
Hb_000612_070 |
0.2122633479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000048_090 |
0.2150325106 |
transcription factor |
TF Family: SET |
hypothetical protein JCGZ_06559 [Jatropha curcas] |
| 11 |
Hb_077562_040 |
0.2285062187 |
- |
- |
hypothetical protein JCGZ_24573 [Jatropha curcas] |
| 12 |
Hb_001488_110 |
0.2373194735 |
- |
- |
- |
| 13 |
Hb_003376_170 |
0.2386700273 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X2 [Jatropha curcas] |
| 14 |
Hb_126398_010 |
0.241169758 |
- |
- |
- |
| 15 |
Hb_002639_070 |
0.2450502121 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 16 |
Hb_000184_120 |
0.2471716147 |
- |
- |
hypothetical protein JCGZ_18526 [Jatropha curcas] |
| 17 |
Hb_002081_100 |
0.2484605477 |
- |
- |
- |
| 18 |
Hb_064201_010 |
0.2552620613 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 19 |
Hb_000340_480 |
0.2563200301 |
- |
- |
- |
| 20 |
Hb_004375_060 |
0.2576063602 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |