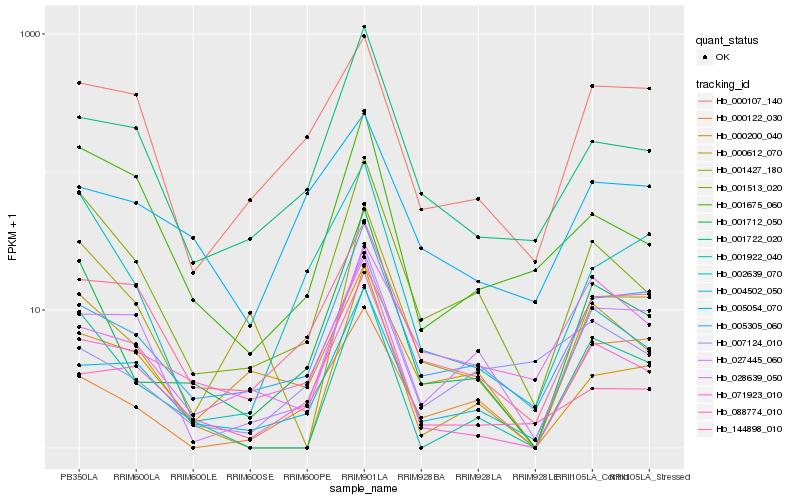
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001712_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 2 |
Hb_001513_020 |
0.1656792446 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 3 |
Hb_005305_060 |
0.1868625417 |
- |
- |
hypothetical protein MIMGU_mgv1a015111mg [Erythranthe guttata] |
| 4 |
Hb_002639_070 |
0.1910223124 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 5 |
Hb_001922_040 |
0.1950639926 |
- |
- |
reticulon family protein [Populus trichocarpa] |
| 6 |
Hb_071923_010 |
0.2117543875 |
- |
- |
- |
| 7 |
Hb_000612_070 |
0.2152799956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001722_020 |
0.2175126155 |
- |
- |
- |
| 9 |
Hb_144898_010 |
0.2272827681 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 10 |
Hb_001427_180 |
0.2347654281 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 11 |
Hb_007124_010 |
0.2438384464 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_027445_060 |
0.245008301 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_004502_050 |
0.2488268643 |
- |
- |
hypothetical protein POPTR_1488s00200g [Populus trichocarpa] |
| 14 |
Hb_000122_030 |
0.2514490903 |
- |
- |
PREDICTED: uncharacterized protein LOC104606081, partial [Nelumbo nucifera] |
| 15 |
Hb_005054_070 |
0.2537974981 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001675_060 |
0.2545711402 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 17 |
Hb_000107_140 |
0.2565969518 |
- |
- |
- |
| 18 |
Hb_088774_010 |
0.2569209593 |
- |
- |
- |
| 19 |
Hb_000200_040 |
0.2578290566 |
- |
- |
- |
| 20 |
Hb_028639_050 |
0.2581307694 |
- |
- |
Anopheles gambiae str. PEST AGAP012660-PA [Anopheles gambiae str. PEST] |