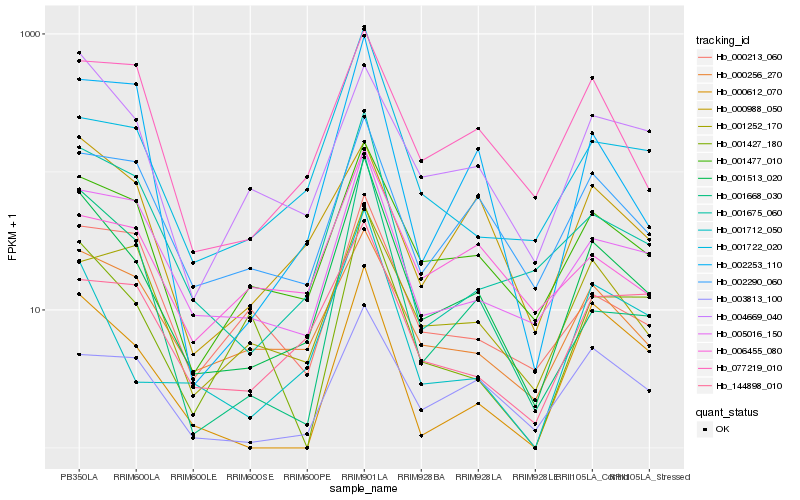
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001513_020 |
0.0 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 2 |
Hb_001477_010 |
0.1431791274 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 13-like [Cicer arietinum] |
| 3 |
Hb_001675_060 |
0.157194266 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 4 |
Hb_005016_150 |
0.1619562104 |
- |
- |
- |
| 5 |
Hb_001712_050 |
0.1656792446 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 6 |
Hb_000256_270 |
0.1660039851 |
- |
- |
PREDICTED: large proline-rich protein bag6-B isoform X1 [Jatropha curcas] |
| 7 |
Hb_001668_030 |
0.1734806785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003813_100 |
0.1773088641 |
- |
- |
PREDICTED: proteasome subunit beta type-3-A [Cucumis sativus] |
| 9 |
Hb_002290_060 |
0.1797929578 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 10 |
Hb_002253_110 |
0.1826068289 |
- |
- |
cysteine desulfurylase, putative [Ricinus communis] |
| 11 |
Hb_004669_040 |
0.185115084 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 12 |
Hb_001722_020 |
0.1861994547 |
- |
- |
- |
| 13 |
Hb_077219_010 |
0.1879673126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_144898_010 |
0.1925937327 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 15 |
Hb_001427_180 |
0.1930403753 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 16 |
Hb_000988_050 |
0.1969857676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006455_080 |
0.1970795918 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 18 |
Hb_000612_070 |
0.1975030751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000213_060 |
0.1987003334 |
- |
- |
PREDICTED: RRP12-like protein isoform X1 [Jatropha curcas] |
| 20 |
Hb_001252_170 |
0.1990655555 |
desease resistance |
Gene Name: Exostosin |
hypothetical protein PRUPE_ppa001357mg [Prunus persica] |