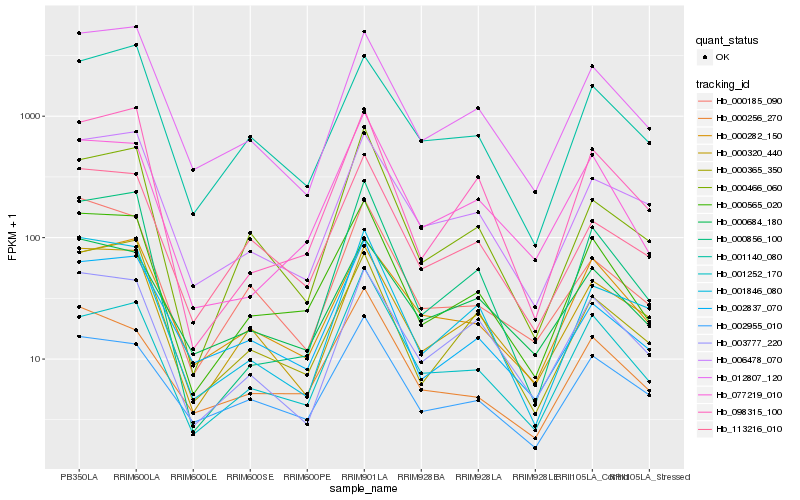
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000565_020 |
0.0 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 2 |
Hb_077219_010 |
0.0989077456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_113216_010 |
0.1056059698 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 4 |
Hb_012807_120 |
0.1061155723 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 5 |
Hb_002837_070 |
0.1095203383 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_098315_100 |
0.1130382433 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001140_080 |
0.1131302586 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 8 |
Hb_000256_270 |
0.1131909269 |
- |
- |
PREDICTED: large proline-rich protein bag6-B isoform X1 [Jatropha curcas] |
| 9 |
Hb_000320_440 |
0.1148727364 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 2/3/4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000856_100 |
0.1151563457 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 11 |
Hb_000282_150 |
0.116811227 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 12 |
Hb_002955_010 |
0.116877406 |
- |
- |
PREDICTED: Werner syndrome ATP-dependent helicase homolog [Jatropha curcas] |
| 13 |
Hb_000466_060 |
0.1187820612 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 14 |
Hb_000185_090 |
0.1190902518 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 15 |
Hb_001252_170 |
0.1191487763 |
desease resistance |
Gene Name: Exostosin |
hypothetical protein PRUPE_ppa001357mg [Prunus persica] |
| 16 |
Hb_000365_350 |
0.1200975926 |
- |
- |
PREDICTED: uncharacterized protein LOC105649068 [Jatropha curcas] |
| 17 |
Hb_003777_220 |
0.1204206681 |
transcription factor |
TF Family: SET |
PREDICTED: protein SET DOMAIN GROUP 41 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006478_070 |
0.1204900173 |
- |
- |
PREDICTED: uncharacterized protein LOC105649957 [Jatropha curcas] |
| 19 |
Hb_001846_080 |
0.122784126 |
- |
- |
PREDICTED: poly(rC)-binding protein 3-like [Nicotiana sylvestris] |
| 20 |
Hb_000684_180 |
0.1235946346 |
- |
- |
PREDICTED: importin subunit alpha-9 isoform X1 [Jatropha curcas] |