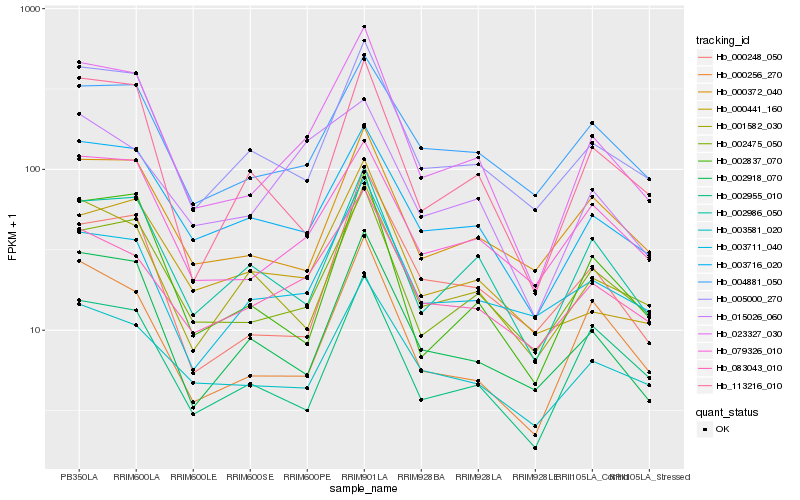
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023327_030 |
0.0 |
- |
- |
osmotin-like protein [Hevea brasiliensis] |
| 2 |
Hb_002475_050 |
0.1106950064 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 3 |
Hb_005000_270 |
0.1134192878 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000256_270 |
0.1144779801 |
- |
- |
PREDICTED: large proline-rich protein bag6-B isoform X1 [Jatropha curcas] |
| 5 |
Hb_079326_010 |
0.1190273239 |
- |
- |
PREDICTED: uncharacterized protein LOC105630719 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002918_070 |
0.1224330786 |
- |
- |
hypothetical protein POPTR_0011s05530g [Populus trichocarpa] |
| 7 |
Hb_003716_020 |
0.1236453484 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |
| 8 |
Hb_083043_010 |
0.1252510565 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 9 |
Hb_002837_070 |
0.1275696623 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002986_050 |
0.128447918 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 11 |
Hb_003581_020 |
0.1310751852 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 12 |
Hb_113216_010 |
0.1323757486 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 13 |
Hb_000248_050 |
0.1359006874 |
- |
- |
PREDICTED: uncharacterized protein LOC105647198 isoform X3 [Jatropha curcas] |
| 14 |
Hb_002955_010 |
0.13746487 |
- |
- |
PREDICTED: Werner syndrome ATP-dependent helicase homolog [Jatropha curcas] |
| 15 |
Hb_000441_160 |
0.137535659 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 16 |
Hb_000372_040 |
0.1386086704 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_015026_060 |
0.1391255354 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 18 |
Hb_001582_030 |
0.1400876567 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 19 |
Hb_004881_050 |
0.1401673132 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 20 |
Hb_003711_040 |
0.140207351 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |