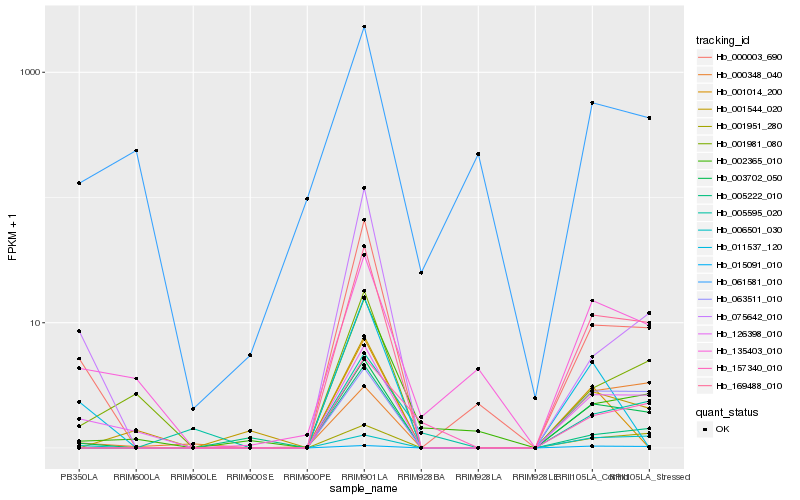
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003702_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649649 [Jatropha curcas] |
| 2 |
Hb_169488_010 |
0.087663699 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_063511_010 |
0.1777829905 |
- |
- |
DNA-directed RNA polymerase II subunit RPB4 [Gossypium arboreum] |
| 4 |
Hb_001951_280 |
0.1790566767 |
- |
- |
- |
| 5 |
Hb_000003_690 |
0.1824579838 |
- |
- |
- |
| 6 |
Hb_126398_010 |
0.1898664498 |
- |
- |
- |
| 7 |
Hb_015091_010 |
0.2093194985 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 8 |
Hb_001544_020 |
0.2158187507 |
- |
- |
- |
| 9 |
Hb_001981_080 |
0.2309191999 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 isoform X4 [Musa acuminata subsp. malaccensis] |
| 10 |
Hb_157340_010 |
0.2451924712 |
- |
- |
hypothetical protein CISIN_1g0109951mg, partial [Citrus sinensis] |
| 11 |
Hb_002365_010 |
0.2535399547 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 12 |
Hb_005595_020 |
0.256710955 |
- |
- |
- |
| 13 |
Hb_135403_010 |
0.2575447595 |
- |
- |
HEV1.1 [Hevea brasiliensis] |
| 14 |
Hb_005222_010 |
0.2582870238 |
- |
- |
hypothetical protein MTR_1g033950 [Medicago truncatula] |
| 15 |
Hb_006501_030 |
0.2643298338 |
- |
- |
hypothetical protein POPTR_0006s17720g [Populus trichocarpa] |
| 16 |
Hb_075642_010 |
0.2722324016 |
- |
- |
PREDICTED: uncharacterized protein LOC105647831 [Jatropha curcas] |
| 17 |
Hb_061581_010 |
0.286383327 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_001014_200 |
0.291341847 |
- |
- |
- |
| 19 |
Hb_000348_040 |
0.2955394487 |
- |
- |
- |
| 20 |
Hb_011537_120 |
0.2974315572 |
- |
- |
- |