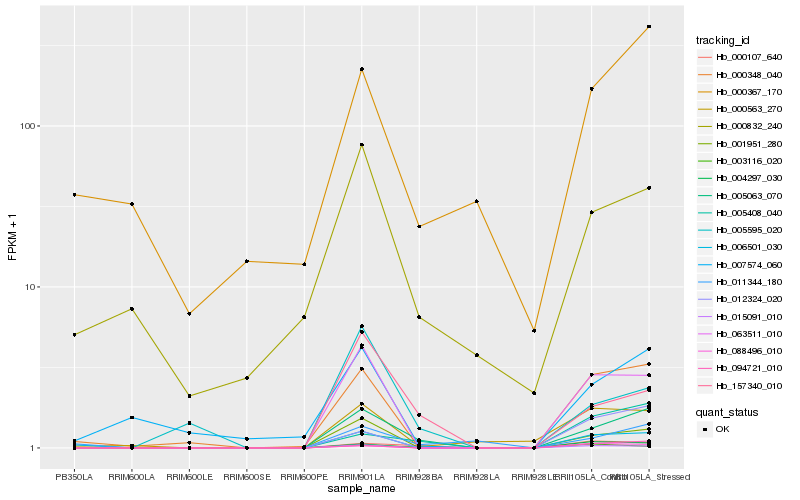
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005063_070 |
0.0 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 2 |
Hb_001951_280 |
0.2044351383 |
- |
- |
- |
| 3 |
Hb_063511_010 |
0.2265787134 |
- |
- |
DNA-directed RNA polymerase II subunit RPB4 [Gossypium arboreum] |
| 4 |
Hb_157340_010 |
0.2275425613 |
- |
- |
hypothetical protein CISIN_1g0109951mg, partial [Citrus sinensis] |
| 5 |
Hb_000107_640 |
0.2282432445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_015091_010 |
0.2446605883 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 7 |
Hb_000348_040 |
0.2461442094 |
- |
- |
- |
| 8 |
Hb_006501_030 |
0.2673078095 |
- |
- |
hypothetical protein POPTR_0006s17720g [Populus trichocarpa] |
| 9 |
Hb_005408_040 |
0.2689448762 |
- |
- |
- |
| 10 |
Hb_003116_020 |
0.2793422333 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 11 |
Hb_011344_180 |
0.2793775958 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 12 |
Hb_004297_030 |
0.2848998389 |
- |
- |
PREDICTED: uncharacterized protein LOC105633198 [Jatropha curcas] |
| 13 |
Hb_005595_020 |
0.2860121056 |
- |
- |
- |
| 14 |
Hb_094721_010 |
0.2908185597 |
- |
- |
PREDICTED: uncharacterized protein LOC104809818, partial [Tarenaya hassleriana] |
| 15 |
Hb_000832_240 |
0.2912338221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012324_020 |
0.2931951824 |
- |
- |
- |
| 17 |
Hb_000563_270 |
0.294697229 |
- |
- |
- |
| 18 |
Hb_088496_010 |
0.3005988588 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_007574_060 |
0.3009399528 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 27-like [Jatropha curcas] |
| 20 |
Hb_000367_170 |
0.3017594998 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |