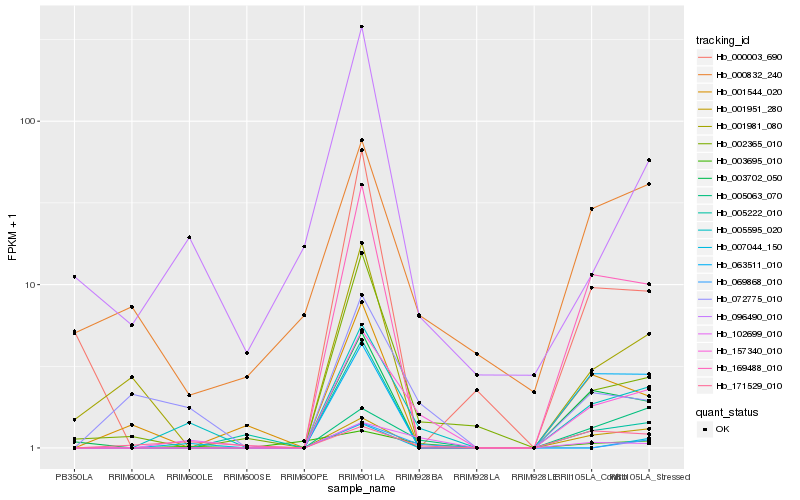
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005595_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_157340_010 |
0.1836072678 |
- |
- |
hypothetical protein CISIN_1g0109951mg, partial [Citrus sinensis] |
| 3 |
Hb_169488_010 |
0.2364568183 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic-like [Populus euphratica] |
| 4 |
Hb_072775_010 |
0.2524578082 |
- |
- |
60S ribosomal L37-3 -like protein [Gossypium arboreum] |
| 5 |
Hb_003702_050 |
0.256710955 |
- |
- |
PREDICTED: uncharacterized protein LOC105649649 [Jatropha curcas] |
| 6 |
Hb_001951_280 |
0.2657700585 |
- |
- |
- |
| 7 |
Hb_002365_010 |
0.2700965583 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 8 |
Hb_096490_010 |
0.2838405899 |
- |
- |
hypothetical protein CICLE_v100210772mg, partial [Citrus clementina] |
| 9 |
Hb_005063_070 |
0.2860121056 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 10 |
Hb_102699_010 |
0.2864141343 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR type disease resistance protein [Populus trichocarpa] |
| 11 |
Hb_063511_010 |
0.2888835835 |
- |
- |
DNA-directed RNA polymerase II subunit RPB4 [Gossypium arboreum] |
| 12 |
Hb_007044_150 |
0.2928418683 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 8A-like [Jatropha curcas] |
| 13 |
Hb_171529_010 |
0.2984872963 |
- |
- |
- |
| 14 |
Hb_069868_010 |
0.303866462 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 15 |
Hb_000003_690 |
0.3080485961 |
- |
- |
- |
| 16 |
Hb_005222_010 |
0.3082850806 |
- |
- |
hypothetical protein MTR_1g033950 [Medicago truncatula] |
| 17 |
Hb_000832_240 |
0.3095419715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001981_080 |
0.3097280203 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 isoform X4 [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_001544_020 |
0.312825483 |
- |
- |
- |
| 20 |
Hb_003695_010 |
0.3180633478 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |