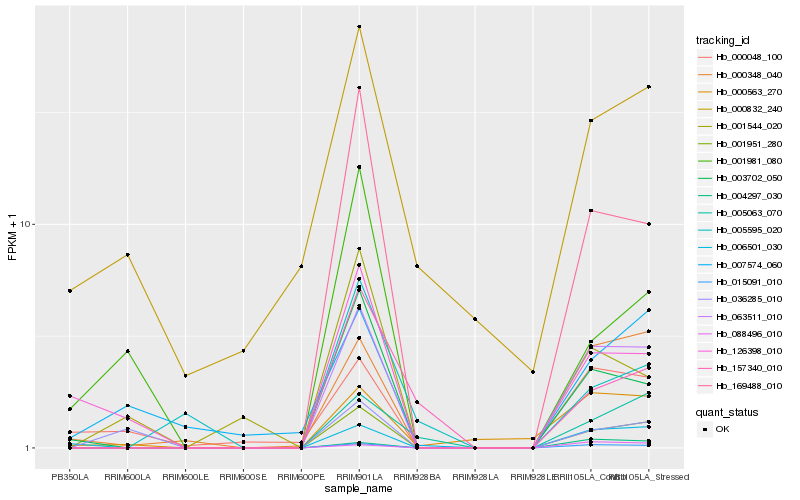
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_280 |
0.0 |
- |
- |
- |
| 2 |
Hb_063511_010 |
0.0756935864 |
- |
- |
DNA-directed RNA polymerase II subunit RPB4 [Gossypium arboreum] |
| 3 |
Hb_015091_010 |
0.1268898807 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 4 |
Hb_169488_010 |
0.1598837584 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_003702_050 |
0.1790566767 |
- |
- |
PREDICTED: uncharacterized protein LOC105649649 [Jatropha curcas] |
| 6 |
Hb_000348_040 |
0.2021831213 |
- |
- |
- |
| 7 |
Hb_005063_070 |
0.2044351383 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 8 |
Hb_006501_030 |
0.2125564377 |
- |
- |
hypothetical protein POPTR_0006s17720g [Populus trichocarpa] |
| 9 |
Hb_004297_030 |
0.2505727698 |
- |
- |
PREDICTED: uncharacterized protein LOC105633198 [Jatropha curcas] |
| 10 |
Hb_157340_010 |
0.251131919 |
- |
- |
hypothetical protein CISIN_1g0109951mg, partial [Citrus sinensis] |
| 11 |
Hb_126398_010 |
0.2623890557 |
- |
- |
- |
| 12 |
Hb_005595_020 |
0.2657700585 |
- |
- |
- |
| 13 |
Hb_000563_270 |
0.2712909648 |
- |
- |
- |
| 14 |
Hb_088496_010 |
0.2773131216 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_001544_020 |
0.2804573535 |
- |
- |
- |
| 16 |
Hb_000048_100 |
0.2816842737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007574_060 |
0.2836380644 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 27-like [Jatropha curcas] |
| 18 |
Hb_001981_080 |
0.2908546402 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 isoform X4 [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_036285_010 |
0.2937938593 |
- |
- |
- |
| 20 |
Hb_000832_240 |
0.295397756 |
- |
- |
conserved hypothetical protein [Ricinus communis] |