| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
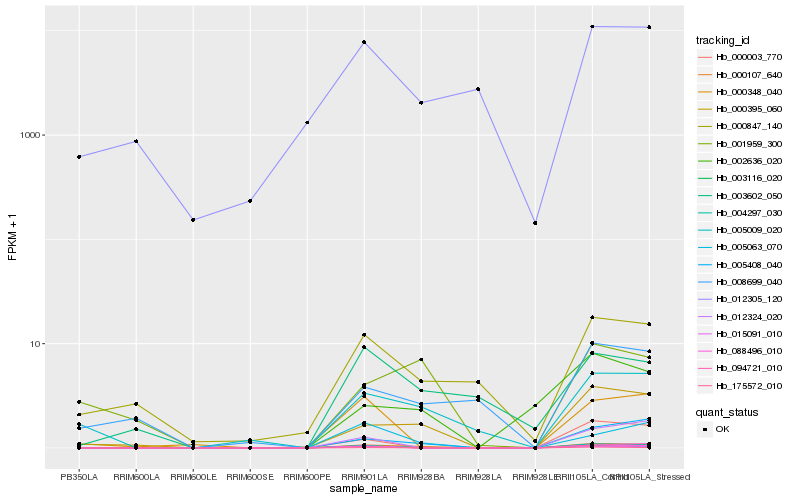
Hb_000107_640 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_175572_010 |
0.1335685385 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 3 |
Hb_003116_020 |
0.1523538622 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 4 |
Hb_000395_060 |
0.1932861805 |
- |
- |
- |
| 5 |
Hb_005408_040 |
0.2088593998 |
- |
- |
- |
| 6 |
Hb_005063_070 |
0.2282432445 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 7 |
Hb_005009_020 |
0.2335150456 |
- |
- |
- |
| 8 |
Hb_004297_030 |
0.2472126307 |
- |
- |
PREDICTED: uncharacterized protein LOC105633198 [Jatropha curcas] |
| 9 |
Hb_088496_010 |
0.2502478632 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_003602_050 |
0.2693361725 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000847_140 |
0.2695940674 |
- |
- |
- |
| 12 |
Hb_002636_020 |
0.2756427124 |
- |
- |
- |
| 13 |
Hb_001959_300 |
0.282730737 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase large subunit [Jatropha curcas] |
| 14 |
Hb_015091_010 |
0.2830697153 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 15 |
Hb_000003_770 |
0.2881476234 |
- |
- |
- |
| 16 |
Hb_000348_040 |
0.2940045779 |
- |
- |
- |
| 17 |
Hb_094721_010 |
0.294082967 |
- |
- |
PREDICTED: uncharacterized protein LOC104809818, partial [Tarenaya hassleriana] |
| 18 |
Hb_012324_020 |
0.2950917017 |
- |
- |
- |
| 19 |
Hb_008699_040 |
0.3024327032 |
- |
- |
- |
| 20 |
Hb_012305_120 |
0.3045222297 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |