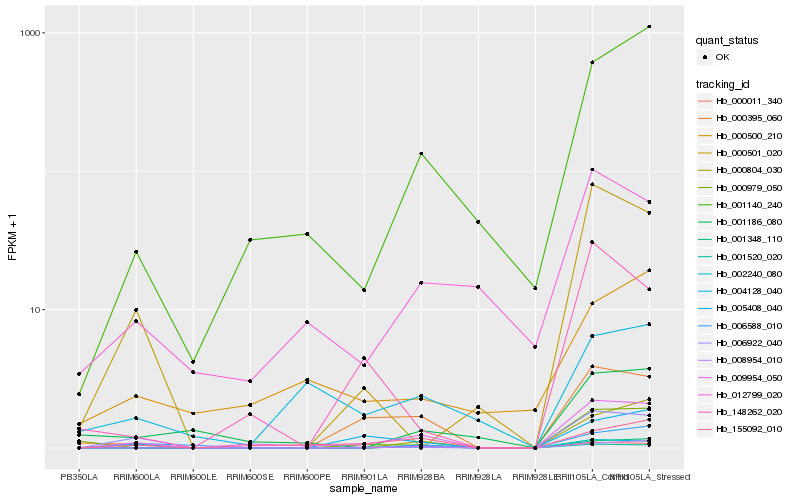
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009954_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104611877 isoform X2 [Nelumbo nucifera] |
| 2 |
Hb_155092_010 |
0.1879221742 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 3 |
Hb_001140_240 |
0.2013205058 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_000395_060 |
0.2274794285 |
- |
- |
- |
| 5 |
Hb_001348_110 |
0.2288152511 |
- |
- |
PREDICTED: esterase-like [Jatropha curcas] |
| 6 |
Hb_001520_020 |
0.2298960606 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 7 |
Hb_000011_340 |
0.2305147574 |
- |
- |
PREDICTED: protein LURP-one-related 11-like [Jatropha curcas] |
| 8 |
Hb_008954_010 |
0.2425693044 |
- |
- |
- |
| 9 |
Hb_006922_040 |
0.2592413043 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000804_030 |
0.259778612 |
- |
- |
Protein C14orf111, putative [Ricinus communis] |
| 11 |
Hb_012799_020 |
0.2605078785 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_002240_080 |
0.2691774082 |
- |
- |
- |
| 13 |
Hb_001186_080 |
0.2726383073 |
- |
- |
PREDICTED: F-box protein FBW2-like [Nelumbo nucifera] |
| 14 |
Hb_000500_210 |
0.2746339901 |
- |
- |
- |
| 15 |
Hb_006588_010 |
0.2793990639 |
- |
- |
- |
| 16 |
Hb_005408_040 |
0.2797067539 |
- |
- |
- |
| 17 |
Hb_000501_020 |
0.2797568291 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 18 |
Hb_000979_050 |
0.2821733776 |
- |
- |
- |
| 19 |
Hb_004128_040 |
0.2823424347 |
- |
- |
- |
| 20 |
Hb_148262_020 |
0.2829218017 |
- |
- |
- |