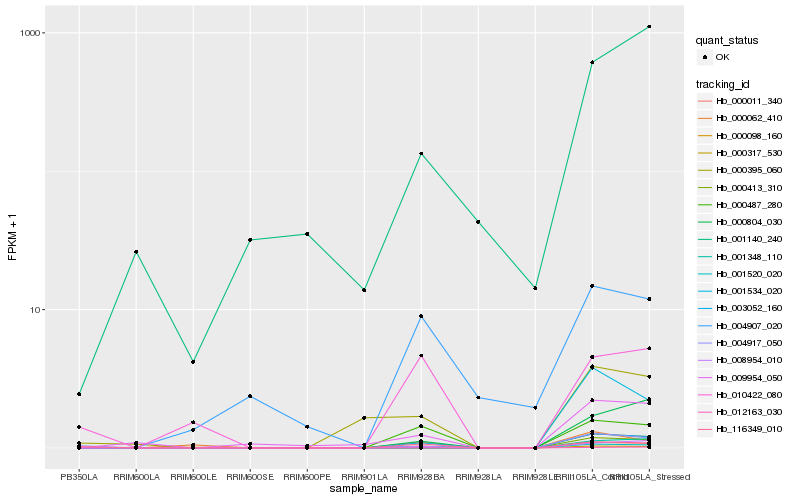
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_340 |
0.0 |
- |
- |
PREDICTED: protein LURP-one-related 11-like [Jatropha curcas] |
| 2 |
Hb_001520_020 |
0.0019765903 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 3 |
Hb_001348_110 |
0.1370202861 |
- |
- |
PREDICTED: esterase-like [Jatropha curcas] |
| 4 |
Hb_000487_280 |
0.1440302651 |
- |
- |
- |
| 5 |
Hb_008954_010 |
0.1458924871 |
- |
- |
- |
| 6 |
Hb_000804_030 |
0.2100118326 |
- |
- |
Protein C14orf111, putative [Ricinus communis] |
| 7 |
Hb_116349_010 |
0.2296434677 |
- |
- |
PREDICTED: uncharacterized protein LOC105763153 [Gossypium raimondii] |
| 8 |
Hb_009954_050 |
0.2305147574 |
- |
- |
PREDICTED: uncharacterized protein LOC104611877 isoform X2 [Nelumbo nucifera] |
| 9 |
Hb_001534_020 |
0.2394838113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000062_410 |
0.249717566 |
- |
- |
PREDICTED: uncharacterized protein LOC105644766 [Jatropha curcas] |
| 11 |
Hb_000395_060 |
0.2542345942 |
- |
- |
- |
| 12 |
Hb_004907_020 |
0.2665061181 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 13 |
Hb_010422_080 |
0.269041213 |
- |
- |
- |
| 14 |
Hb_004917_050 |
0.2693471349 |
- |
- |
- |
| 15 |
Hb_012163_030 |
0.2708975371 |
- |
- |
PREDICTED: uncharacterized protein LOC105647861 [Jatropha curcas] |
| 16 |
Hb_000317_530 |
0.2782926011 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 17 |
Hb_001140_240 |
0.2814409461 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_000098_160 |
0.2845819668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003052_160 |
0.2845819668 |
- |
- |
- |
| 20 |
Hb_000413_310 |
0.2845819673 |
- |
- |
- |