| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
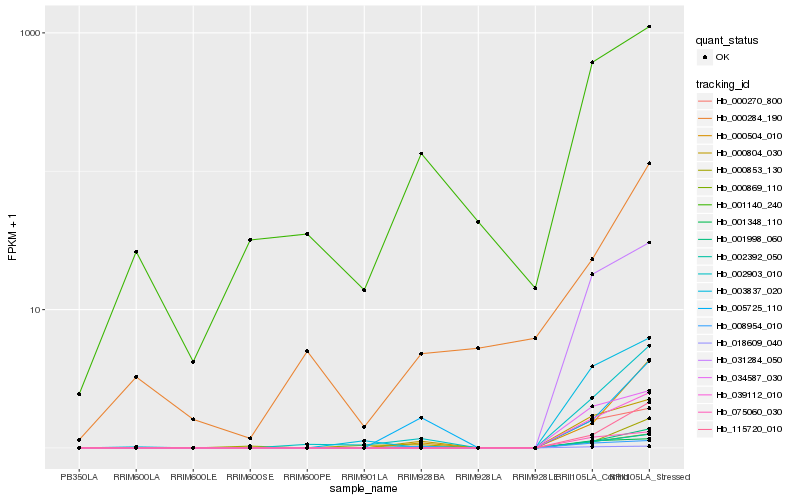
Hb_002903_010 |
0.0 |
- |
- |
60S ribosomal protein L37-3 [Morus notabilis] |
| 2 |
Hb_000504_010 |
0.1423192292 |
- |
- |
oleosin 18.2 kDa-like [Jatropha curcas] |
| 3 |
Hb_001998_060 |
0.1543696432 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_115720_010 |
0.1619837497 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 5 |
Hb_039112_010 |
0.1679827126 |
- |
- |
- |
| 6 |
Hb_000804_030 |
0.168653356 |
- |
- |
Protein C14orf111, putative [Ricinus communis] |
| 7 |
Hb_000869_110 |
0.1686761368 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002392_050 |
0.1687597314 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X4 [Jatropha curcas] |
| 9 |
Hb_003837_020 |
0.188968879 |
- |
- |
PREDICTED: uncharacterized protein LOC103710465 isoform X1 [Phoenix dactylifera] |
| 10 |
Hb_031284_050 |
0.2011721514 |
- |
- |
- |
| 11 |
Hb_034587_030 |
0.2119826994 |
- |
- |
- |
| 12 |
Hb_018609_040 |
0.2145927645 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 13 |
Hb_000270_800 |
0.2146814786 |
- |
- |
- |
| 14 |
Hb_075060_030 |
0.2147581343 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 15 |
Hb_005725_110 |
0.2193778254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000284_190 |
0.2225071682 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 17 |
Hb_000853_130 |
0.2234890571 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 18 |
Hb_008954_010 |
0.2245975369 |
- |
- |
- |
| 19 |
Hb_001140_240 |
0.2256048515 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_001348_110 |
0.2313734998 |
- |
- |
PREDICTED: esterase-like [Jatropha curcas] |