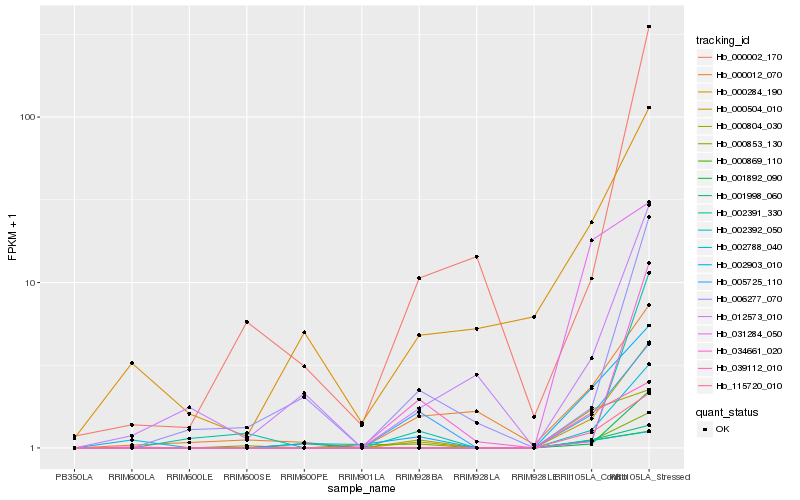
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000504_010 |
0.0 |
- |
- |
oleosin 18.2 kDa-like [Jatropha curcas] |
| 2 |
Hb_115720_010 |
0.1178335316 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 3 |
Hb_002903_010 |
0.1423192292 |
- |
- |
60S ribosomal protein L37-3 [Morus notabilis] |
| 4 |
Hb_001998_060 |
0.160777706 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002788_040 |
0.1849308885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005725_110 |
0.2009532645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_039112_010 |
0.2021542219 |
- |
- |
- |
| 8 |
Hb_000869_110 |
0.2035809551 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_002392_050 |
0.2037512628 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X4 [Jatropha curcas] |
| 10 |
Hb_001892_090 |
0.2257213375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000853_130 |
0.2331696925 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 12 |
Hb_000002_170 |
0.2368936653 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B [Jatropha curcas] |
| 13 |
Hb_000804_030 |
0.2398293843 |
- |
- |
Protein C14orf111, putative [Ricinus communis] |
| 14 |
Hb_000284_190 |
0.2421027563 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 15 |
Hb_002391_330 |
0.2487003021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012573_010 |
0.2506435462 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 17 |
Hb_034661_020 |
0.2507634409 |
- |
- |
hypothetical protein JCGZ_04352 [Jatropha curcas] |
| 18 |
Hb_006277_070 |
0.2558292402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_031284_050 |
0.2561707431 |
- |
- |
- |
| 20 |
Hb_000012_070 |
0.2635126174 |
- |
- |
cytochrome P450, putative [Ricinus communis] |