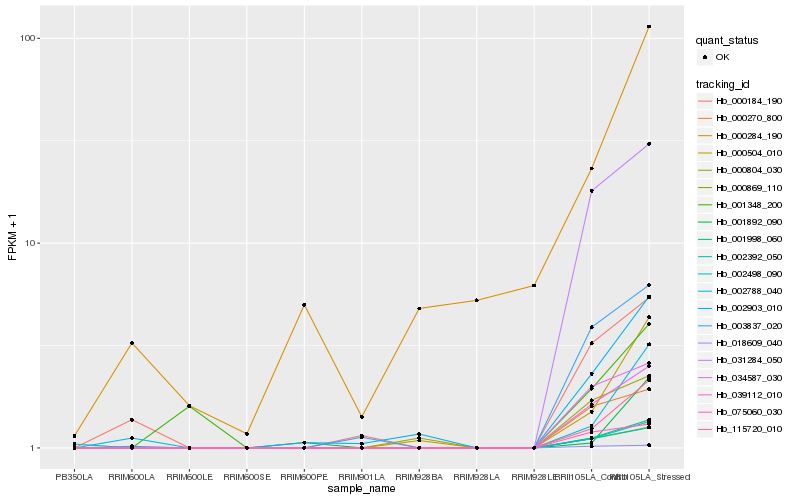
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115720_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 2 |
Hb_001998_060 |
0.0695433105 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000504_010 |
0.1178335316 |
- |
- |
oleosin 18.2 kDa-like [Jatropha curcas] |
| 4 |
Hb_039112_010 |
0.1214353252 |
- |
- |
- |
| 5 |
Hb_000869_110 |
0.1231333675 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_002392_050 |
0.1233357759 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X4 [Jatropha curcas] |
| 7 |
Hb_002903_010 |
0.1619837497 |
- |
- |
60S ribosomal protein L37-3 [Morus notabilis] |
| 8 |
Hb_002788_040 |
0.1688551443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_031284_050 |
0.1835152378 |
- |
- |
- |
| 10 |
Hb_003837_020 |
0.1978303083 |
- |
- |
PREDICTED: uncharacterized protein LOC103710465 isoform X1 [Phoenix dactylifera] |
| 11 |
Hb_034587_030 |
0.1997442898 |
- |
- |
- |
| 12 |
Hb_018609_040 |
0.2035166067 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 13 |
Hb_000270_800 |
0.203643958 |
- |
- |
- |
| 14 |
Hb_075060_030 |
0.2037539543 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 15 |
Hb_002498_090 |
0.2247385201 |
- |
- |
- |
| 16 |
Hb_001892_090 |
0.2364233063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000804_030 |
0.2460089525 |
- |
- |
Protein C14orf111, putative [Ricinus communis] |
| 18 |
Hb_000184_190 |
0.2482829878 |
- |
- |
hypothetical protein VITISV_016970 [Vitis vinifera] |
| 19 |
Hb_000284_190 |
0.2653855082 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 20 |
Hb_001348_200 |
0.2697686317 |
- |
- |
- |