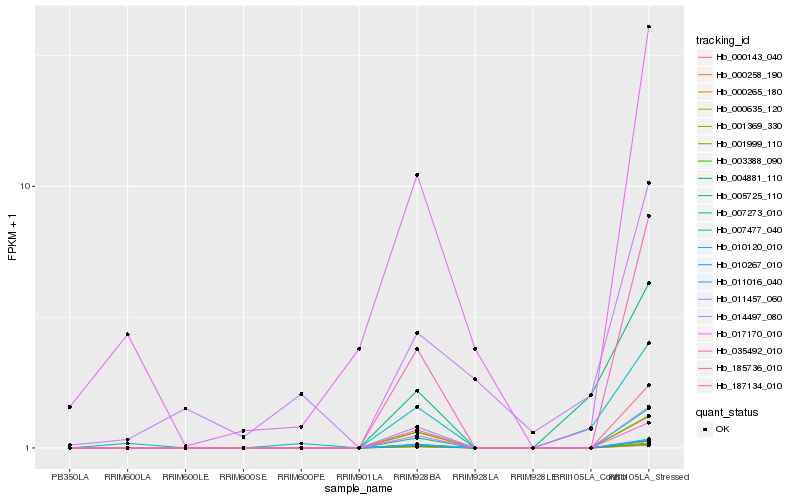
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007477_040 |
0.0 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 2 |
Hb_005725_110 |
0.1567441249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_187134_010 |
0.2563120723 |
- |
- |
- |
| 4 |
Hb_007273_010 |
0.2574690807 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 5 |
Hb_185736_010 |
0.2595206037 |
- |
- |
- |
| 6 |
Hb_011016_040 |
0.2607801489 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000143_040 |
0.2607855057 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 8 |
Hb_000258_190 |
0.2609097933 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_003388_090 |
0.261016737 |
- |
- |
putative beta-D-xylosidase 5 -like protein [Gossypium arboreum] |
| 10 |
Hb_000265_180 |
0.2612108998 |
- |
- |
Alpha/beta-Hydrolases superfamily protein, putative [Theobroma cacao] |
| 11 |
Hb_004881_110 |
0.2616234729 |
- |
- |
- |
| 12 |
Hb_001369_330 |
0.2616403452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010120_010 |
0.2617937117 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 14 |
Hb_010267_010 |
0.2620019195 |
- |
- |
- |
| 15 |
Hb_017170_010 |
0.2661027674 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 16 |
Hb_035492_010 |
0.2676777312 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 17 |
Hb_000635_120 |
0.268774938 |
- |
- |
- |
| 18 |
Hb_001999_110 |
0.2688114154 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 19 |
Hb_014497_080 |
0.2688917668 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 20 |
Hb_011457_060 |
0.2691323159 |
- |
- |
PREDICTED: uncharacterized protein LOC105649957 [Jatropha curcas] |