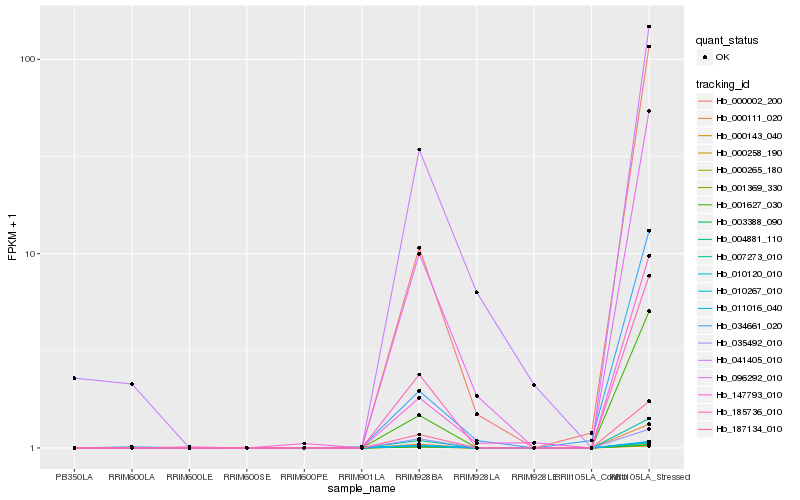
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096292_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_25718 [Jatropha curcas] |
| 2 |
Hb_000111_020 |
0.0872579606 |
- |
- |
hypothetical protein SORBIDRAFT_04g003755 [Sorghum bicolor] |
| 3 |
Hb_185736_010 |
0.0882473386 |
- |
- |
- |
| 4 |
Hb_007273_010 |
0.0929334849 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 5 |
Hb_187134_010 |
0.096891959 |
- |
- |
- |
| 6 |
Hb_001627_030 |
0.0973772417 |
- |
- |
hypothetical protein JCGZ_11752 [Jatropha curcas] |
| 7 |
Hb_000002_200 |
0.1049885148 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF096 [Brassica rapa] |
| 8 |
Hb_041405_010 |
0.1164677184 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Glycine max] |
| 9 |
Hb_147793_010 |
0.1187789937 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PUB24-like [Jatropha curcas] |
| 10 |
Hb_034661_020 |
0.1209786987 |
- |
- |
hypothetical protein JCGZ_04352 [Jatropha curcas] |
| 11 |
Hb_011016_040 |
0.1676920934 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000143_040 |
0.1677131512 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 13 |
Hb_000258_190 |
0.1681997202 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_003388_090 |
0.1686153588 |
- |
- |
putative beta-D-xylosidase 5 -like protein [Gossypium arboreum] |
| 15 |
Hb_000265_180 |
0.1693630202 |
- |
- |
Alpha/beta-Hydrolases superfamily protein, putative [Theobroma cacao] |
| 16 |
Hb_004881_110 |
0.1709233492 |
- |
- |
- |
| 17 |
Hb_001369_330 |
0.1709863718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010120_010 |
0.1715565082 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 19 |
Hb_010267_010 |
0.1723228284 |
- |
- |
- |
| 20 |
Hb_035492_010 |
0.1907022805 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |