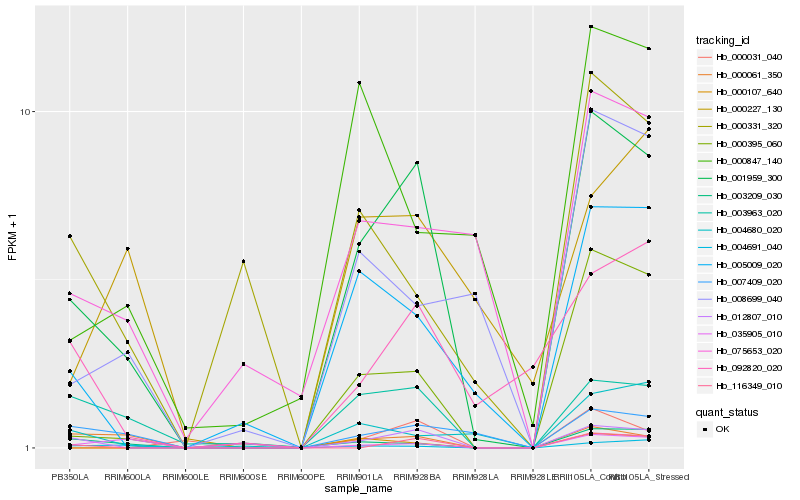
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_300 |
0.0 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase large subunit [Jatropha curcas] |
| 2 |
Hb_000395_060 |
0.2185371166 |
- |
- |
- |
| 3 |
Hb_003963_020 |
0.2209393926 |
- |
- |
PREDICTED: uncharacterized protein LOC100256672 [Vitis vinifera] |
| 4 |
Hb_012807_010 |
0.2292357933 |
- |
- |
- |
| 5 |
Hb_005009_020 |
0.2410246691 |
- |
- |
- |
| 6 |
Hb_007409_020 |
0.2656744788 |
- |
- |
- |
| 7 |
Hb_004691_040 |
0.269287576 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 8 |
Hb_035905_010 |
0.2774497074 |
- |
- |
PREDICTED: uncharacterized protein LOC105627956 [Jatropha curcas] |
| 9 |
Hb_116349_010 |
0.278167704 |
- |
- |
PREDICTED: uncharacterized protein LOC105763153 [Gossypium raimondii] |
| 10 |
Hb_075653_020 |
0.2787837439 |
- |
- |
- |
| 11 |
Hb_008699_040 |
0.2807707768 |
- |
- |
- |
| 12 |
Hb_000107_640 |
0.282730737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_092820_020 |
0.284245373 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000331_320 |
0.2924076558 |
- |
- |
- |
| 15 |
Hb_004680_020 |
0.2938538968 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105789475 [Gossypium raimondii] |
| 16 |
Hb_000847_140 |
0.297508066 |
- |
- |
- |
| 17 |
Hb_003209_030 |
0.3038228645 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 18 |
Hb_000227_130 |
0.3045130252 |
- |
- |
PREDICTED: peroxiredoxin-2F, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000061_350 |
0.3050885779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000031_040 |
0.3085325191 |
- |
- |
centromere protein O [Medicago truncatula] |