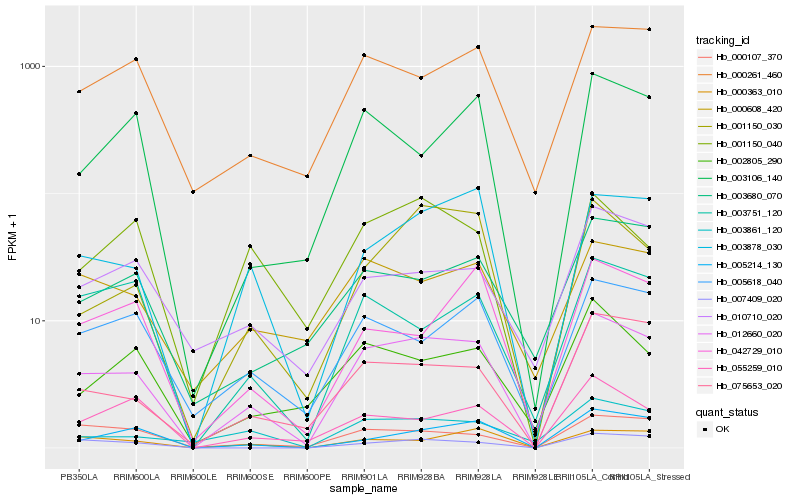
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012660_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_003878_030 |
0.1435364248 |
- |
- |
hypothetical protein POPTR_0011s11120g [Populus trichocarpa] |
| 3 |
Hb_001150_030 |
0.1601469008 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 4 |
Hb_075653_020 |
0.1602814949 |
- |
- |
- |
| 5 |
Hb_000363_010 |
0.1696547497 |
- |
- |
- |
| 6 |
Hb_005618_040 |
0.1714314649 |
- |
- |
- |
| 7 |
Hb_000107_370 |
0.1768124953 |
- |
- |
PREDICTED: uncharacterized protein At2g17340-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000261_460 |
0.1771937451 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 9 |
Hb_005214_130 |
0.179083197 |
- |
- |
- |
| 10 |
Hb_007409_020 |
0.1831759296 |
- |
- |
- |
| 11 |
Hb_003106_140 |
0.1839361934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000608_420 |
0.1861691383 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 13 |
Hb_003861_120 |
0.186737111 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_003751_120 |
0.1891140165 |
- |
- |
- |
| 15 |
Hb_055259_010 |
0.1919709161 |
- |
- |
- |
| 16 |
Hb_003680_070 |
0.1934529171 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002805_290 |
0.1943469704 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_042729_010 |
0.1958287621 |
- |
- |
- |
| 19 |
Hb_010710_020 |
0.196785176 |
- |
- |
PREDICTED: uncharacterized protein LOC103322096 [Prunus mume] |
| 20 |
Hb_001150_040 |
0.1975353971 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |