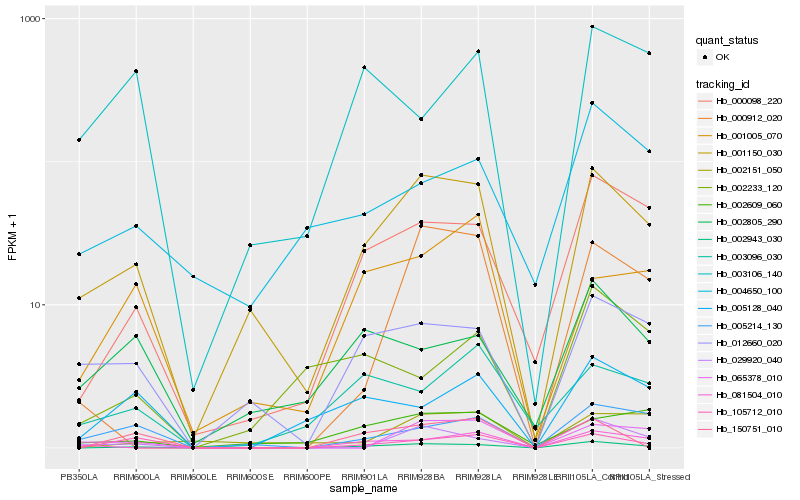
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002943_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000098_220 |
0.1814105302 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_001150_030 |
0.1920013299 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 4 |
Hb_105712_010 |
0.224292771 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 5 |
Hb_012660_020 |
0.2622270373 |
- |
- |
- |
| 6 |
Hb_150751_010 |
0.2679334986 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 7 |
Hb_081504_010 |
0.2700270509 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 8 |
Hb_029920_040 |
0.2712328385 |
- |
- |
PREDICTED: uncharacterized protein LOC105635837 [Jatropha curcas] |
| 9 |
Hb_005128_040 |
0.2775766637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000912_020 |
0.2793069821 |
- |
- |
PREDICTED: uncharacterized protein LOC105645039 [Jatropha curcas] |
| 11 |
Hb_005214_130 |
0.2801666302 |
- |
- |
- |
| 12 |
Hb_002805_290 |
0.2922868918 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_002609_060 |
0.3039982388 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 14 |
Hb_004650_100 |
0.3056615727 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_002151_050 |
0.3065035405 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_002233_120 |
0.3083980272 |
- |
- |
- |
| 17 |
Hb_003106_140 |
0.3097391686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001005_070 |
0.3099544113 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 19 |
Hb_065378_010 |
0.3122547856 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 20 |
Hb_003096_030 |
0.3123191346 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |