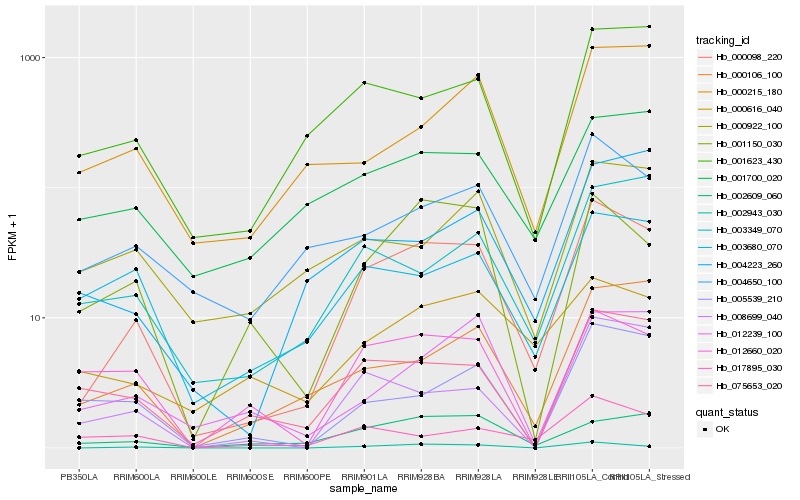
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_220 |
0.0 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 2 |
Hb_002943_030 |
0.1814105302 |
- |
- |
- |
| 3 |
Hb_001623_430 |
0.2050845194 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 4 |
Hb_004650_100 |
0.2072775079 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_075653_020 |
0.209139892 |
- |
- |
- |
| 6 |
Hb_000106_100 |
0.2104879563 |
- |
- |
- |
| 7 |
Hb_002609_060 |
0.2114085734 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 8 |
Hb_001150_030 |
0.2117941478 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 9 |
Hb_008699_040 |
0.2127737711 |
- |
- |
- |
| 10 |
Hb_003349_070 |
0.2159455538 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 11 |
Hb_003680_070 |
0.2191401543 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_017895_030 |
0.2194174179 |
transcription factor |
TF Family: C2H2 |
- |
| 13 |
Hb_012239_100 |
0.2230472967 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 14 |
Hb_001700_020 |
0.2241774296 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 15 |
Hb_000616_040 |
0.2249324646 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 16 |
Hb_005539_210 |
0.2255240332 |
- |
- |
- |
| 17 |
Hb_012660_020 |
0.2263375144 |
- |
- |
- |
| 18 |
Hb_004223_260 |
0.2286305091 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 19 |
Hb_000215_180 |
0.2312663905 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 20 |
Hb_000922_100 |
0.2342893918 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |