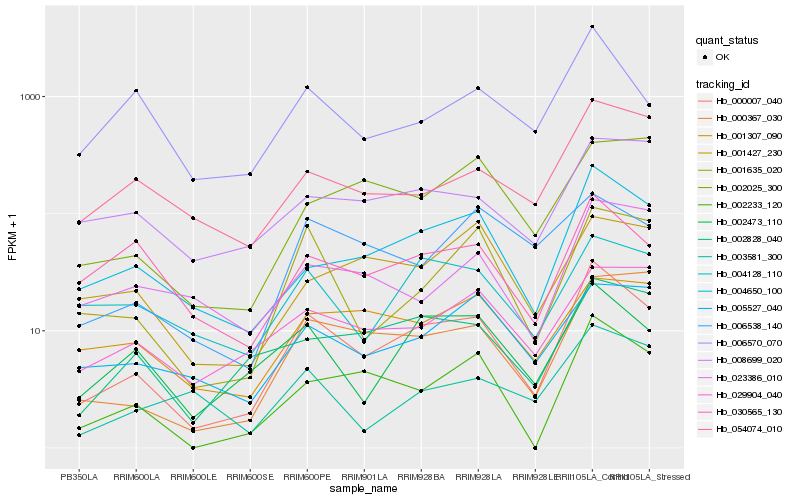
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000007_040 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a017616mg [Erythranthe guttata] |
| 2 |
Hb_004650_100 |
0.1530905958 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_002233_120 |
0.1699897134 |
- |
- |
- |
| 4 |
Hb_002473_110 |
0.175059069 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_002828_040 |
0.1823644724 |
- |
- |
transmembrane protein, putative [Medicago truncatula] |
| 6 |
Hb_001635_020 |
0.1832139482 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 7 |
Hb_000367_030 |
0.1889280104 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 8 |
Hb_006570_070 |
0.1970259067 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 9 |
Hb_029904_040 |
0.200186904 |
- |
- |
putative aspartate aminotransferase protein, partial [Elaeis guineensis] |
| 10 |
Hb_054074_010 |
0.2017506187 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 11 |
Hb_030565_130 |
0.2037160598 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 12 |
Hb_005527_040 |
0.2039739169 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 13 |
Hb_004128_110 |
0.2053645917 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 14 |
Hb_001307_090 |
0.2107657797 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |
| 15 |
Hb_006538_140 |
0.2109476312 |
- |
- |
PREDICTED: uncharacterized protein LOC103341483 [Prunus mume] |
| 16 |
Hb_003581_300 |
0.2145615406 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008699_020 |
0.2151254887 |
- |
- |
eukaryotic translation initiation factor 5A isoform IV [Hevea brasiliensis] |
| 18 |
Hb_023386_010 |
0.2169165165 |
- |
- |
PREDICTED: histidine triad nucleotide-binding protein 3 [Jatropha curcas] |
| 19 |
Hb_002025_300 |
0.2178502797 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 20 |
Hb_001427_230 |
0.2201533969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |