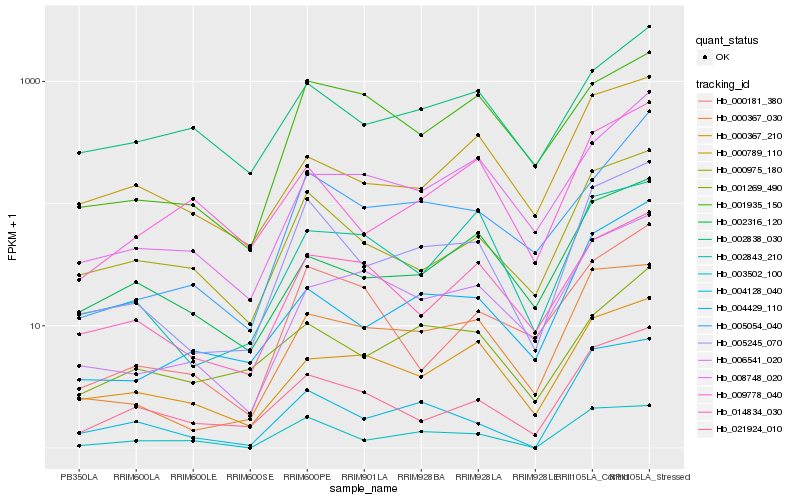
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005245_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 2 |
Hb_000367_030 |
0.1287208331 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 3 |
Hb_000975_180 |
0.1324433363 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 4 |
Hb_003502_100 |
0.1466588837 |
- |
- |
- |
| 5 |
Hb_021924_010 |
0.1486665926 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 6 |
Hb_004429_110 |
0.1552597441 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 7 |
Hb_004128_040 |
0.156078068 |
- |
- |
- |
| 8 |
Hb_005054_040 |
0.1601209848 |
- |
- |
PREDICTED: 60S ribosomal protein L35a-1 [Gossypium raimondii] |
| 9 |
Hb_014834_030 |
0.1655065976 |
- |
- |
- |
| 10 |
Hb_002316_120 |
0.1691403107 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 11 |
Hb_000367_210 |
0.170808607 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 12 |
Hb_008748_020 |
0.1713819383 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 13 |
Hb_002843_210 |
0.171755386 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 14 |
Hb_006541_020 |
0.1729474104 |
- |
- |
unknown [Lotus japonicus] |
| 15 |
Hb_002838_030 |
0.1731442596 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 16 |
Hb_000789_110 |
0.1744371664 |
- |
- |
RecName: Full=Profilin-1; AltName: Full=Pollen allergen Hev b 8.0101; AltName: Allergen=Hev b 8.0101 [Hevea brasiliensis] |
| 17 |
Hb_009778_040 |
0.1756334551 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 18 |
Hb_001269_490 |
0.1772921243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000181_380 |
0.1779254401 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 20 |
Hb_001935_150 |
0.1785575408 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |