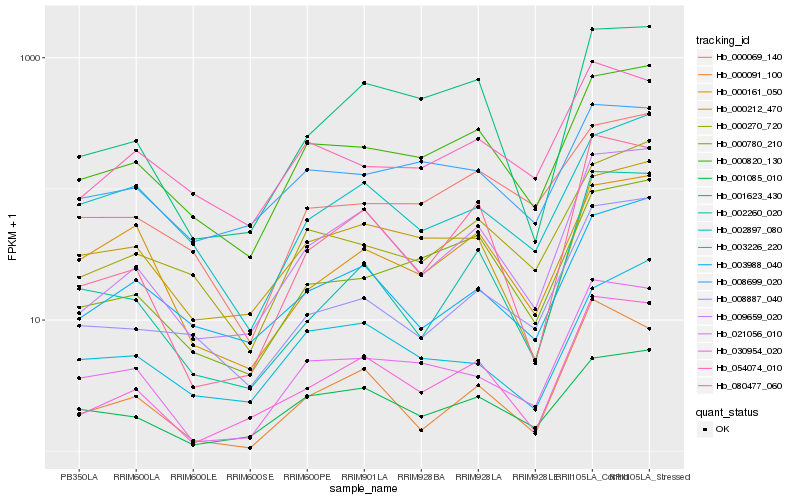
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021056_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000820_130 |
0.1336164545 |
- |
- |
PREDICTED: uncharacterized protein LOC105769309 isoform X1 [Gossypium raimondii] |
| 3 |
Hb_030954_020 |
0.1362469329 |
- |
- |
hypothetical protein RCOM_1330200 [Ricinus communis] |
| 4 |
Hb_009659_020 |
0.1447703601 |
- |
- |
PREDICTED: uncharacterized protein LOC103870090 [Brassica rapa] |
| 5 |
Hb_000212_470 |
0.1467989868 |
- |
- |
hypothetical protein CICLE_v10033256mg [Citrus clementina] |
| 6 |
Hb_001085_010 |
0.1500178608 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 4 isoform X3 [Populus euphratica] |
| 7 |
Hb_000780_210 |
0.1516393846 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 8 |
Hb_001623_430 |
0.1554537068 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 9 |
Hb_008699_020 |
0.1572256152 |
- |
- |
eukaryotic translation initiation factor 5A isoform IV [Hevea brasiliensis] |
| 10 |
Hb_054074_010 |
0.1577543512 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 11 |
Hb_000091_100 |
0.1593706614 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_080477_060 |
0.1600839645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008887_040 |
0.161896076 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_003226_220 |
0.1621854424 |
- |
- |
hypothetical protein CICLE_v10022836mg [Citrus clementina] |
| 15 |
Hb_002897_080 |
0.1702833438 |
- |
- |
hypothetical protein JCGZ_03306 [Jatropha curcas] |
| 16 |
Hb_000161_050 |
0.1710503301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003988_040 |
0.171783919 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 18 |
Hb_000270_720 |
0.171784285 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 19 |
Hb_002260_020 |
0.1725482629 |
- |
- |
- |
| 20 |
Hb_000069_140 |
0.1726936892 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |