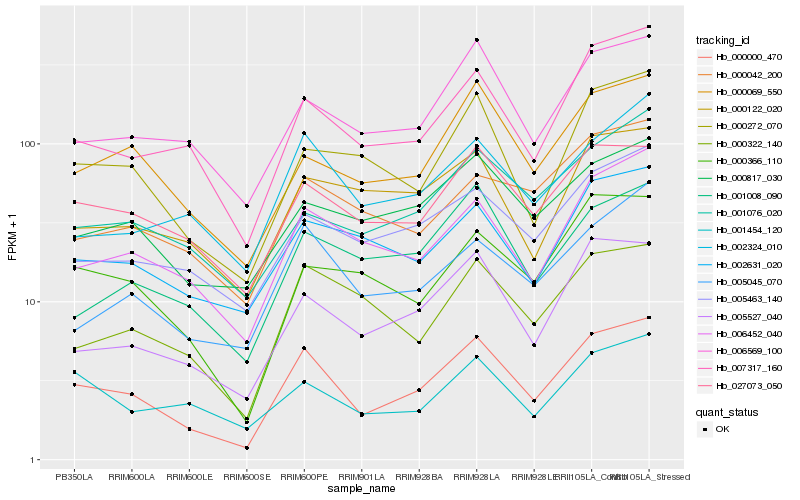
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_470 |
0.0 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X4 [Jatropha curcas] |
| 2 |
Hb_007317_160 |
0.1158690735 |
- |
- |
PREDICTED: uncharacterized protein At2g27730, mitochondrial [Jatropha curcas] |
| 3 |
Hb_005527_040 |
0.1159128281 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 4 |
Hb_006452_040 |
0.1205313578 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |
| 5 |
Hb_027073_050 |
0.123062391 |
- |
- |
PREDICTED: uncharacterized protein LOC105635813 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000069_550 |
0.1247920335 |
- |
- |
PREDICTED: uncharacterized protein LOC104596457 isoform X1 [Nelumbo nucifera] |
| 7 |
Hb_006569_100 |
0.1250534446 |
- |
- |
hypothetical protein POPTR_0005s26990g [Populus trichocarpa] |
| 8 |
Hb_000272_070 |
0.1270529871 |
- |
- |
snare protein YKt [Hevea brasiliensis] |
| 9 |
Hb_005463_140 |
0.1312322521 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18 [Jatropha curcas] |
| 10 |
Hb_005045_070 |
0.1317585963 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 11 |
Hb_000817_030 |
0.1323549842 |
- |
- |
mak, putative [Ricinus communis] |
| 12 |
Hb_002631_020 |
0.1332386557 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 13 |
Hb_000322_140 |
0.1341918605 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 14 |
Hb_000366_110 |
0.1353513718 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |
| 15 |
Hb_002324_010 |
0.137615602 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 16 |
Hb_001008_090 |
0.1392584753 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 17 |
Hb_000122_020 |
0.1408276057 |
- |
- |
PREDICTED: alpha-soluble NSF attachment protein 2 [Jatropha curcas] |
| 18 |
Hb_001076_020 |
0.1443138558 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 19 |
Hb_001454_120 |
0.1443869905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000042_200 |
0.1448698305 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X1 [Vitis vinifera] |