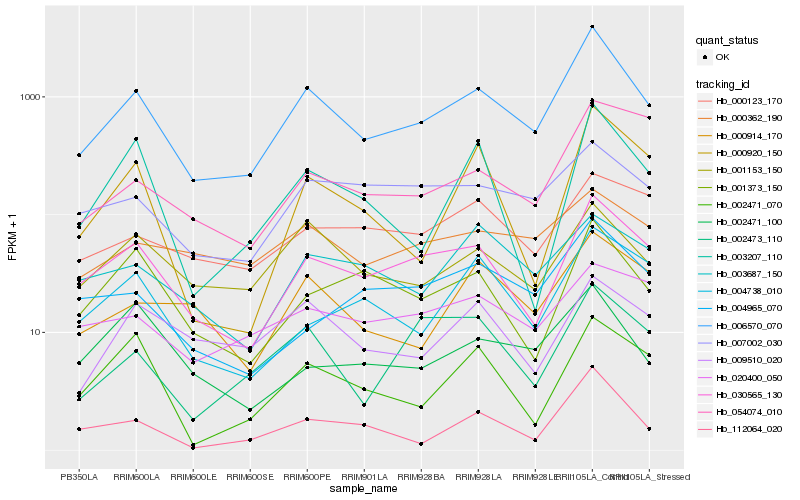
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006570_070 |
0.0 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 2 |
Hb_030565_130 |
0.1265933744 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 3 |
Hb_001153_150 |
0.1615576634 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 4 |
Hb_112064_020 |
0.1647977346 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 5 |
Hb_000914_170 |
0.1738885128 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 6 |
Hb_001373_150 |
0.1788248926 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 7 |
Hb_002473_110 |
0.181642318 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_007002_030 |
0.1817157502 |
- |
- |
PREDICTED: transmembrane protein 205 [Jatropha curcas] |
| 9 |
Hb_003207_110 |
0.1840032715 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl coenzyme A synthase [Hevea brasiliensis] |
| 10 |
Hb_009510_020 |
0.1860052572 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 11 |
Hb_004965_070 |
0.1871650096 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 12 |
Hb_004738_010 |
0.1876853022 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 13 |
Hb_002471_070 |
0.1896042016 |
- |
- |
Tartrate-resistant acid phosphatase type 5 precursor, putative [Ricinus communis] |
| 14 |
Hb_003687_150 |
0.1899080823 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002471_100 |
0.189991062 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000920_150 |
0.1905002804 |
- |
- |
annexin [Manihot esculenta] |
| 17 |
Hb_000362_190 |
0.1908922638 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 18 |
Hb_000123_170 |
0.1924987492 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 19 |
Hb_020400_050 |
0.1935297143 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 20 |
Hb_054074_010 |
0.1936069489 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |